Abstract
Background: Muse (multilineage-differentiating stress-enduring) cells are a pluripotent subpopulation of mesenchymal stem cells (MSCs), characterized by their stress tolerance and significant potential in regenerative medicine. However, their low abundance poses challenges for isolation. This study aims to evaluate the efficacy of severe stress conditions—including low temperature, severe hypoxia, and collagenase treatment (LHC)—in isolating Muse cells.
Methods: Human umbilical cord-derived MSCs (hUCMSCs) were treated with 0.1% collagenase D in DMEM at 37 °C for 30 minutes, then incubated at 4 °C for 16 hours in sealed containers completely filled with the collagenase-containing medium. Muse cell enrichment following this treatment was quantified by flow cytometry. Morphological characteristics of the Muse-enriched-cell (MEC) populations were examined under adherent and suspension culture conditions. Their trilineage differentiation potential into adipocytes (mesoderm), hepatocyte-like cells (endoderm), and neuron-like cells (ectoderm) was evaluated. Additionally, the expression of pluripotency-associated genes (Sox2, Nanog, and Oct4) was assessed via RT-qPCR, and chromosomal stability was confirmed through G-banding karyotype analysis.
Results: The percentage of SSEA-3+ cells in MEC populations (53.47 ± 17.16%) was significantly higher than in native hUCMSCs (3.43 ± 1.50%). MECs formed clusters resembling embryonic stem cells in suspension culture and differentiated into adipocytes (lipid droplet+), hepatocyte-like cells (cytokeratin-7+), and neuron-like cells (MAP-2+). MEC-SC populations exhibited significantly higher mRNA expression of Nanog, Sox2, and Oct4 compared to MEC-AC populations and native hUCMSCs.
Conclusion: The LHC method provides a promising and efficient approach for isolating Muse cells, which could significantly advance their applications in regenerative medicine.
Introduction
In recent decades, mesenchymal stem cells (MSCs) have been extensively studied and applied in regenerative medicine due to their remarkable therapeutic potential. These cells exhibit essential properties that facilitate tissue repair, including immunomodulatory effects, secretion of growth factors, and the ability to differentiate into various functional cell types. These characteristics make MSCs promising candidates for treating various diseases, including osteoarthritis, diabetes, and limb ischemia1.
MSCs can be isolated from multiple tissue sources, such as adipose tissue, bone marrow, and umbilical cord-derived tissues1. Interestingly, MSCs derived from different sources exhibit distinct biological properties. For example, umbilical cord-derived MSCs display strong immunomodulatory effects, bone marrow-derived MSCs provide superior regenerative support, and adipose tissue-derived MSCs possess an enhanced capacity for extracellular matrix production2, 3. Moreover, even within the same tissue source, MSC populations exhibit heterogeneity in their characteristics and functional capacities. This cellular heterogeneity is well-documented and is thought to influence the therapeutic efficacy of MSC-based therapies1, 4.
In 2010, a distinct subpopulation of MSCs, known as multilineage-differentiating stress-enduring (Muse) cells, was first identified by a research group at Tohoku University in Japan4. These cells express the stage-specific embryonic antigen-3 (SSEA-3), a marker typically associated with embryonic development. Muse cells exhibit several pluripotent-like characteristics, including self-renewal, expression of key pluripotency markers (such as Nanog, Sox2, and Oct3/4), and the capacity to differentiate into cells of all three germ layers. Unlike traditional pluripotent stem cells, however, Muse cells do not form teratomas when injected into animals, highlighting their safety for therapeutic applications. Additionally, they possess unique biological properties, such as high resistance to stress conditions, efficient migration to injury sites, and an intrinsic ability to detect and repair DNA damage5.
Muse cells have been identified in various tissues and organs, including bone marrow, peripheral blood, adipose tissue, skin, umbilical cord tissue, spleen, trachea, pancreas, and the amniotic membrane5, 6. Rather than being confined to a specific niche, Muse cells are thought to be sparsely distributed throughout the body. Muse cells constitute approximately 0.01 to 0.03 percent of the mononuclear cell fraction in bone marrow, while in peripheral blood, their proportion ranges from 0.01 to 0.2 percent. Their presence in peripheral blood is believed to result from migration from other tissues, such as bone marrow or the spleen, and their abundance may fluctuate in response to the body's physiological state5.
Muse cells have been successfully isolated from a range of human tissues, including skin fibroblasts, adipose tissue, bone marrow, and the amniotic membrane4, 5, 6, 7. They have also been isolated from mesenchymal stem cell populations in the bone marrow, adipose tissue, and skin of several animal species, including rabbits8, mice9, rats10, pigs11, sheep12, goats13, and dogs14. Most studies have focused on isolating Muse cells from adipose tissue and bone marrow. Within mesenchymal stem cell populations, Muse cells typically comprise 0.5 to 3 percent of the total population, with variations depending on the source tissue or species.
Several methods have been developed to isolate or enrich Muse cells, which can be broadly categorized into two groups: cell sorting techniques (such as fluorescence-activated cell sorting [FACS] and magnetic-activated cell sorting [MACS]) and stress condition treatments4.
FACS is one of the most effective methods for isolating Muse cells. In this method, cells are labeled with antibodies targeting specific surface markers, such as SSEA-3, a critical marker for Muse cells. Depending on the cell source or culture stage, SSEA-3 may be used alone or combined with other markers (e.g., CD105 for MSCs or CD45 for blood cells)15, 16. After incubation with the antibodies, the labeled cells are sorted using a flow cytometry-based system. FACS offers high purity and precision but requires sophisticated equipment and extensive technical expertise, and it may also affect cell viability.
Similar to FACS, MACS relies on cell-surface markers to isolate Muse cells. Cells are labeled with antibodies conjugated to magnetic beads and passed through a magnetic column. The bead-bound cells are retained, while non-target cells flow through. Although MACS is simpler and less costly than FACS, it has lower precision and purity.
The stress condition treatment (SCT) method is a simple approach for isolating and enriching Muse cells. This technique utilizes commonly available and inexpensive chemicals, making it accessible without costly equipment. In SCT, Muse cells can be isolated or enriched from tissue or cultured cells. Cells in culture are exposed to stress conditions, such as nutrient deprivation, serum starvation, or prolonged trypsin exposure. However, these techniques generally yield a relatively low proportion of Muse cells, with SSEA-3-positive cells comprising about 8 to 11 percent4.
Some studies have explored the use of combined stress conditions to isolate Muse cells directly from adipose tissue. In these cases, the adipose tissue is treated with collagenase and then incubated at 4°C for 16 hours in sealed containers filled with collagenase solution, creating a low-oxygen, closed environment. This method has yielded a cell population with a high percentage of SSEA-3-positive cells, ranging from 57 to 90 percent12, 17, 18. However, this approach has primarily been applied to freshly isolated tissues. Based on this, we hypothesize that a combination of collagenase treatment, low temperature, and limited gas exchange (hereafter referred to as LHC conditions) may also be effective for enriching Muse cells from secondary cell cultures. This method is expected to selectively eliminate non-Muse cells, thereby increasing the proportion of Muse cells within the population. Accordingly, this study investigates the percentage of SSEA-3-positive cells in human umbilical cord-derived mesenchymal stem cell cultures following LHC treatment.
Methods
Collagenase Treatment
Human umbilical cord mesenchymal stem cells (hUCMSCs) at passage 4 or 5 were obtained from the Cell Bank of the Stem Cell Institute, University of Science, VNUHCM, Viet Nam. At 70-80% confluence, the cells were harvested using 0.25% Trypsin-EDTA (Gibco, Thermo Fisher Scientific, USA). The cell pellets were washed twice with PBS (300 × g, 5 min) and then incubated in 2 mL of DMEM (Gibco, Thermo Fisher Scientific, USA) containing 0.1% collagenase D (Roche, Merck, USA) at 37°C for 30 minutes. Following incubation, the cells were transferred to a sealed 2 mL cryotube pre-filled with collagenase solution and incubated at 4°C for 16 hours under low-oxygen conditions created by the closed system17. Afterward, the cells were cultured in T25 flasks under adherent conditions for 6–12 hours to remove dead cells, and the candidate Muse-enriched cell (MEC) populations were harvested17.
Flow Cytometry Analysis for MSC Surface Markers
Cells harvested at passages 4–5 were washed twice with phosphate-buffered saline (PBS) and incubated for 15 minutes at 37°C in the dark with the following fluorochrome-conjugated antibodies: Anti-human CD14-FITC, CD34-FITC, CD45-APC, and HLA-DR-FITC (hematopoietic lineage markers), Anti-human CD44-PE, CD73-APC, CD105-PE, and CD90-PE (mesenchymal stromal cell markers). Post-incubation, cells were washed to remove unbound antibodies and analyzed using a BD FACS Melody™ flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Data acquisition was performed with BD FACSuite™ software, and subsequent analysis was conducted using FlowJo™ software (Tree Star, Ashland, OR, USA).
SSEA-3 Expression Analysis
The percentage of SSEA-3 expression in MEC populations was measured after low-oxygen and collagenase (LHC) treatment using flow cytometry (FACSCalibur, BD Biosciences, USA). Briefly, MECs were washed with PBS and suspended in 100 µL of PBS. The cells were then incubated with Alexa Fluor® 488-conjugated SSEA-3 antibody (Invitrogen, Thermo Fisher Scientific, USA) or without the antibody (negative control) at 4°C for 30 minutes. After incubation, the cells were washed twice with PBS and resuspended in 200 µL of PBS. The percentage of SSEA-3-positive cells was quantified using FACSCalibur and analyzed with FlowJo software. Additionally, MECs seeded in 96-well plates were incubated with SSEA-3 antibody, and expression was visualized using an Axio A1 microscope (Carl Zeiss, Germany).
Cluster Information Assay
To prevent adhesion, the flask surface was coated with 1% agarose. MECs were seeded at 20,000 cells per 2 mL in a 6-well plate or T25 flask. The morphology and size of the resulting clusters were measured on day 7.
Adipocyte Differentiation
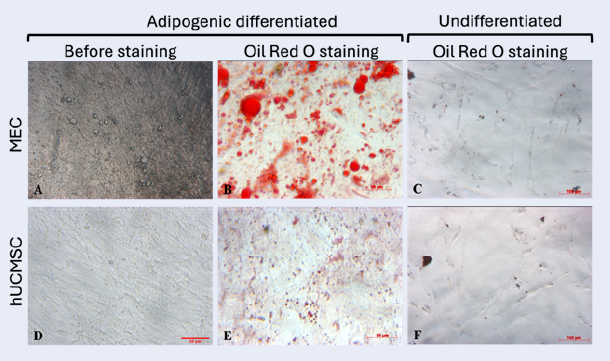
MECs were seeded at 2,500 cells/well in a 96-well plate and cultured in DMEM/F12 (Gibco, Thermo Fisher Scientific, USA) supplemented with 10% FBS (Gibco, Thermo Fisher Scientific, USA) at 37°C with 5% CO2. After 12–24 hours, the medium was replaced with adipocyte differentiation medium (Gibco, Thermo Fisher Scientific, USA), refreshed every 48 hours for 21 days. Lipid droplets were stained with 0.5% Oil Red O (Sigma-Aldrich, USA) prepared as a 3:2 (v/v) solution in distilled water. Oil Red O-positive area was quantified using ImageJ.
Hepatocyte Differentiation
MECs were treated with hepatocyte differentiation medium (DMEM + 10% FBS, 1× ITS (Gibco, Thermo Fisher Scientific, USA), 10 nM dexamethasone, 100 ng/mL HGF (PeproTech, USA), and 50 ng/mL FGF-4 (PeproTech, USA)) for 7 days. Hepatocytes were identified by immunostaining for cytokeratin 7 (CK-7; Abcam, UK). Fluorescence intensity was quantified using ImageJ.
Neuron Differentiation
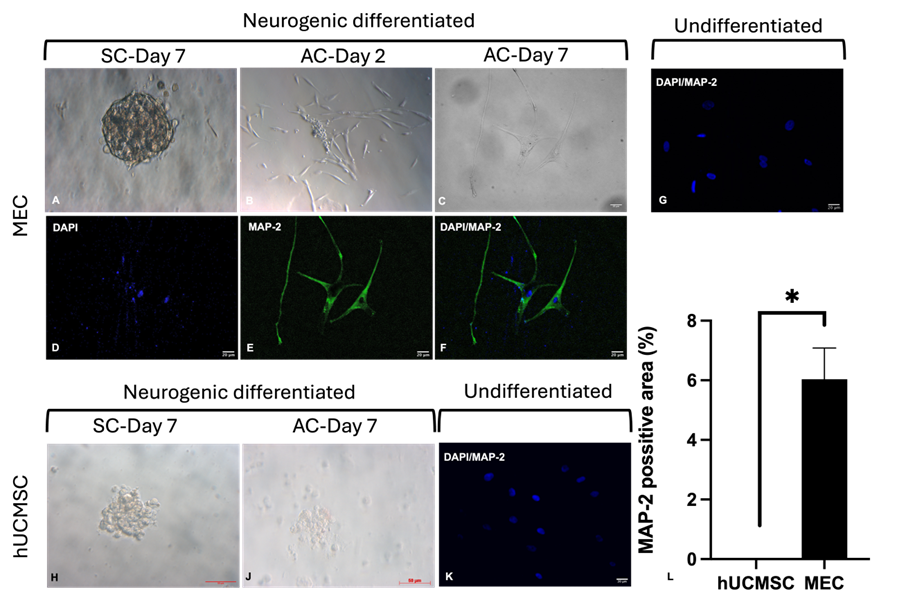
MECs were cultured in suspension in 6-well agarose-coated plates for 7 days in neural differentiation medium A (Neurobasal medium (Gibco, Thermo Fisher Scientific, USA) + 1× B-27, 2 mM L-glutamine, 30 ng/mL bFGF (Miltenyi Biotech, Germany), and 30 ng/mL EGF (PeproTech, USA)). Clusters were then transferred to a 96-well plate and cultured for 7 more days in neural differentiation medium B (DMEM + 2% FBS, 25 ng/mL bFGF, 25 ng/mL BDNF (PeproTech, USA)). Neurons were identified by MAP-2 immunostaining (Thermo Fisher Scientific, USA).
Immunocytochemistry
Cells were fixed in 2% paraformaldehyde (20 min, RT), permeabilized with 0.1% Triton X-100 (Merck, Germany), and blocked with 4% goat serum (Gibco, Thermo Fisher Scientific, USA) + 1% BSA (Bomeibio, China). Primary antibodies (CK-7 for hepatocytes or MAP-2 for neurons) were applied overnight (4°C), followed by secondary antibodies (1 h, RT) and 1 µg/mL DAPI (Merck, Germany) nuclear staining. Images were captured using an Axio A1 microscope (Carl Zeiss, Germany; 20× objective).
RT-qPCR Analysis
Total RNA was extracted from MECs and hUCMSCs using the easy-BLUE™ Total RNA Extraction Kit (iNtRON Biotechnology, Korea). mRNA levels of Nanog, Sox2, and Oct4 were assessed by one-step RT-qPCR (Luna® Universal Kit, New England Biolabs, USA) and quantified via the 2−ΔΔCt method19. Primer sequences are listed inTable 1 20.
Karyotype Analysis
MECs at 70–80% confluency were treated with 0.1 µg/mL Colcemid (Gibco, Thermo Fisher Scientific, USA) for 15 min, incubated in 0.075 M KCl (15 min, 37°C), and fixed in Carnoy’s solution (3:1 methanol:acetic acid). Cells were dropped onto chilled slides, air-dried, treated with 0.25% Trypsin-EDTA (30 sec), and stained with Giemsa stain (Gibco, Thermo Fisher Scientific, USA). Karyotypes were analyzed using Ikaros software (MetaSystems, Germany).
Statistical Analysis
Data are presented as mean ± SD (n = 5). Differences were analyzed by one-way ANOVA with Tukey’s post-hoc test (GraphPad Prism 9; p < 0.05).
| Primer | Sequences | Gene ID |
| Nanog | F: 5’ TAGCAATGGTGTGACGCAGAAG 3’ | NM_024865.2 |
| R: 5’ TCTGGTTGCTCCACATTGGAAGG 3’ | ||
| Sox-2 | F: 5’ CATCACCCACAGCAAATGACAGC 3’ | NM_002701.4 |
| R: 5’ TTGCGTGAGTGTGGATGGGATTG 3’ | ||
| Oct-4 | F: 5’ GAGGCAACCTGGAGAATTTGTTCC 3’ | NM_003106.2 |
| R: 5’ ATGTGGCTGATCTGCTGCAGTG 3’ | ||
| ACTB | F: 5’ GGCGGACTATGACTTAGTTGCGTTACACC 3’ | NM_001101.5 |
| R: 5’AAGTCCTCGGCCACATTGTGAACTTTG 3’ |
Results
Expression of human umbilical cord mesenchymal stem cell markers
Flow cytometry analysis demonstrated that the cultured cells displayed a surface marker profile characteristic of mesenchymal stem cells (MSCs). The cells showed negligible expression (< 2%) of hematopoietic and immune lineage markers (CD14, CD34, CD45, and HLA-DR) but exhibited strong positivity (> 95%) for the canonical MSC markers CD44, CD73, CD90, and CD105 (Figure 1).
Morphology and SSEA-3 expression of MECs
Under adherent culture conditions (MEC-AC), MECs displayed a spindle-shaped morphology, similar to hUCMSCs, and expressed SSEA-3, whereas hUCMSCs exhibited minimal SSEA-3 positivity (Figure 2A,B,D,F). In suspension culture conditions (MEC-SC), MECs formed clusters ranging in size from 90.18 µm to 104.94 µm by day 7. Although hUCMSCs also formed clusters, these were fewer in number (Figure 2C,F). Following LHC treatment, flow cytometry analysis revealed a significantly higher proportion of SSEA-3-positive cells in MEC populations (53.47 ± 17.16%) compared to native hUCMSCs (3.43 ± 1.50%) (p < 0.05, n = 4) (Figure 2G,H).
Differentiation of MECs to adipocytes
MECs were induced to differentiate into adipocytes over 21 days. During this period, they underwent morphological changes, including cytoplasmic enlargement and lipid droplet formation, which progressively increased in size and stained positive with Oil Red O (ORO) dye (Figure 3A,B). Similarly, hUCMSCs also formed lipid droplets and stained ORO-positive (Figure 3D,E). Uninduced cells in both groups lacked lipid droplets and were ORO-negative (Figure 3C,F). The ORO staining area in the MEC group (37.992 ± 3.286%) was significantly larger than in the hUCMSC group (5.110 ± 0.853%) (p < 0.05, n = 3) (Figure 3G).
Differentiation of MECs to hepatocytes
MECs cultured in hepatogenic medium demonstrated endodermal differentiation potential, transitioning from a spindle-shaped to a polygonal morphology after 7 days. These cells were CK-7-positive, indicating hepatocyte differentiation (Figure 4A–C). In contrast, hUCMSCs showed limited CK-7 expression (Figure 4E–G). Uninduced cells in both groups were CK-7-negative (Figure 4D,H). Although CK-7 staining in MECs (14.949 ± 2.199%) was higher than in hUCMSCs (6.455 ± 6.623%), the difference was not statistically significant (n = 3, Figure 4J).
Differentiation of MECs to neurons
To assess neural differentiation potential, MECs were cultured in suspension for 7 days and then transitioned to adherent conditions for another 7 days. During suspension culture, MECs formed compact clusters resembling M-clusters. After transitioning to adherent conditions, the cells spread outward, initially displaying a mesenchymal morphology but later elongating and forming branched processes resembling neurons (Figure 5A–C). Immunocytochemistry confirmed MAP-2 expression in these cells (Figure 5D–F). In contrast, hUCMSCs rarely formed M-clusters in suspension and exhibited poor attachment and limited neuron-like morphology after transitioning to adherent culture (Figure 5H,J). Uninduced cells in both groups were MAP-2-negative (Figure 5G,K). The MAP-2-positive area in MECs (6.038 ± 1.048%) was significantly larger than in hUCMSCs (p < 0.05, n = 3) (Figure 5L).
Expression of pluripotent genes NANOG, SOX-2, and OCT-4
Pluripotent gene expression in MECs was evaluated under adherent (MEC-AC) and suspension (MEC-SC) culture conditions. MEC-SC showed significantly higher mRNA expression levels of NANOG (4.65 ± 0.39-fold), SOX-2 (4.04 ± 0.18-fold), and OCT-4 (2.04 ± 0.12-fold) compared to MEC-AC (1.95 ± 0.08-fold, 1.82 ± 0.13-fold, and 1.52 ± 0.08-fold, respectively) and hUCMSCs (p < 0.05, n = 3) (Figure 6).
Karyotyping
Karyotype analysis of MECs following collagenase treatment revealed a normal karyotype (Figure 7C,D), consistent with that of hUCMSCs (Figure 7A,B).
Discussion
Muse cells are a unique subpopulation within mesenchymal stem cells (MSCs) and are reportedly pluripotent. They exhibit key pluripotency characteristics, including self-renewal, the ability to differentiate into cell types from all three germ layers, and expression of pluripotency markers such as Sox2, Nanog, Oct4, and SSEA-3. Additionally, Muse cells demonstrate remarkable stress tolerance and efficient homing to injury sites, making them a promising therapeutic option for regenerative medicine5. However, their low abundance in tissues and cultured MSC populations presents significant challenges for their isolation and application.
Isolation techniques such as fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS) are commonly used but are costly and technically complex. Stress-induced isolation methods have also been explored but remain limited in efficiency. Recent studies suggest that combining collagenase treatment, hypoxia, and low-temperature (LHC) conditions could yield Muse cells with high purity. However, these studies have predominantly focused on isolating Muse cells directly from tissues12, 17, 18. This study evaluates the efficacy of the LHC method in isolating Muse cells from secondary cell cultures.
Our findings demonstrate that the LHC method is a promising strategy for isolating or enriching Muse cells from human umbilical cord-derived mesenchymal stem cells (hUCMSCs). Following LHC treatment, the proportion of SSEA-3-positive cells in the Muse-enriched cell (MEC) population was significantly higher compared to untreated hUCMSCs. Under adherent culture conditions, MECs retained a fibroblast-like morphology similar to that of hUCMSCs, while in suspension, they formed characteristic cell clusters. The enriched SSEA-3+ cell population also exhibited the ability to differentiate into cell types representative of all three germ layers, including hepatocyte-like and neuron-like cells. Although the observed differentiation efficiencies—14.95 ± 2.20 % for CK-7+ hepatocyte-like cells and 6.04 ± 1.05 % for MAP-2+ neuron-like cells—were modest, these results support the multipotent nature of the enriched Muse cells and validate the potential of the LHC method as a foundational enrichment strategy.
It is important to note that these results represent an initial step toward the development of a Muse cell-based platform from hUCMSCs. The differentiation protocols employed in this study were not optimized for maximal efficiency or functional maturation but served to establish proof of concept for the responsiveness of the enriched cells to lineage-specific cues. Future studies will focus on refining induction conditions, improving lineage-specific differentiation efficiencies, assessing functional characteristics of derived cells, and evaluating therapeutic potential in relevant in vivo models. These steps are essential before considering clinical translation.
Muse cells were originally isolated from human bone marrow MSCs via long-term trypsin incubation. However, this method yielded low efficiency, with only 11.6 % SSEA-3-positive cells in MSCs and 8.6 % in fibroblasts. Alternative stress-induced conditions, such as serum-free culture, HBSS incubation, or low oxygen environments, have also been explored, but these studies lacked quantitative assessments of Muse cell proportions post-treatment4. FACS and MACS have proven more effective, with MACS achieving SSEA-3-positive cell proportions ranging from 63.4 % to 96.29 %21, 22, 23, 24, 25. In our study, the LHC method resulted in approximately 50 % SSEA-3-positive cells, higher than long-term trypsin methods but lower than MACS. These results align with previous studies isolating Muse cells from adipose tissue using collagenase12, 18. However, the proportion of SSEA-3-positive cells in the MEC population varied in our study. This variability may be attributed to differences in donor-derived cell sources and passage numbers. As Muse cells represent a rare subpopulation within MSCs, their frequency is inherently low, subject to donor variability, and may fluctuate throughout the culture process25, 26. Previous studies have reported a wide range in the percentage of Muse cells following collagenase treatment of adipose tissue, with values ranging from approximately 90 %17 to 73.4 ± 18 %12 and 57.7 ± 11.8 %18. Additionally, the relatively small sample size in our study (n = 3–4) represents a limitation. Future studies with larger sample sizes and standardized conditions are needed to confirm these findings. Despite these limitations, the LHC method demonstrates potential as an effective strategy for isolating Muse cells from cultured hUCMSCs.
A defining characteristic of Muse cells is their ability to differentiate into cells from all three germ layers, setting them apart from somatic stem cells. In vitro, Muse cells can differentiate spontaneously or in response to cytokine cocktails into various cell types, including hepatocytes, cholangiocytes (endodermal), neurons, melanocytes, keratinocytes (ectodermal), adipocytes, osteocytes, cardiomyocytes, and glomerular cells (mesodermal). Similarly, in vivo, Muse cells have been shown to differentiate into neuronal cells (ectoderm), hepatocytes (endoderm), and cardiomyocytes and glomerular cells (mesoderm)5.
In this study, MECs demonstrated differentiation into adipocytes, hepatocytes, and neuronal cells. Additionally, MECs exhibited significantly higher expression levels of pluripotency genes, such as Nanog, Sox2, and Oct4, compared to hUCMSC populations. Notably, MECs cultured under suspension conditions expressed these genes at higher levels than those in adherent culture. These findings are consistent with recent studies on Muse cells12, 27, 28. While GAPDH is a commonly used housekeeping gene, several studies have reported its variable expression under specific conditions, including hypoxia. In contrast, ACTB (β-actin) has demonstrated stable expression under hypoxic conditions29, 30, 31. Given the hypoxic conditions in our experimental design, we selected ACTB as the reference gene to ensure accurate normalization and minimize variability in gene expression analysis.
Muse cells are known for their exceptional stress tolerance, attributed to their ability to secrete stress resistance factors such as Serpins and 14-3-3 proteins. Serpins inhibit proteases, including trypsin, thrombin, and neutrophil elastase, while 14-3-3 proteins regulate the cell cycle, DNA repair, and apoptosis resistance5. Muse cells also possess superior DNA repair capabilities through an enhanced non-homologous end joining (NHEJ) mechanism32, 33. This robust DNA repair ability enables Muse cells to survive under conditions lethal to most other stem cells, leaving only Muse cells in the population34.
In our study, severe stress treatments resulted in the death of most non-Muse cells, enriching the surviving population with Muse cells. This survival capacity supports the effectiveness of stress-based methods, such as LHC, in increasing the proportion of Muse cells within a population.
Conclusion
In this study, the LHC method effectively enriched Muse cells within MSC populations. These enriched populations exhibited key characteristics of pluripotent stem cells, including differentiation into cells of all three germ layers and high expression levels of pluripotency-associated genes. Moreover, the LHC method demonstrated advantages in time efficiency and cost-effectiveness, streamlining the Muse cell isolation process. This approach holds significant promise for advancing Muse cell research and enabling their clinical applications.
Abbreviations
ACTB (Beta-actin), ANOVA (Analysis of Variance), BDNF (Brain-Derived Neurotrophic Factor), bFGF (Basic Fibroblast Growth Factor), CD14, CD34, CD44, CD45, CD73, CD90, CD105 (Cluster of Differentiation 14, 34, 44, 45, 73, 90, 105), CK-7 (Cytokeratin 7), DMEM (Dulbecco’s Modified Eagle Medium), EGF (Epidermal Growth Factor), FACS (Fluorescence-Activated Cell Sorting), FBS (Fetal Bovine Serum), FGF-4 (Fibroblast Growth Factor-4), GAPDH (Glyceraldehyde 3-Phosphate Dehydrogenase), HBSS (Hank’s Balanced Salt Solution), HGF (Hepatocyte Growth Factor), HLA-DR (Human Leukocyte Antigen–DR), hUCMSCs (Human Umbilical Cord-Derived Mesenchymal Stem Cells), ITS (Insulin-Transferrin-Selenium), LHC (Low temperature, Hypoxia, and Collagenase treatment), MACS (Magnetic-Activated Cell Sorting), MAP-2 (Microtubule-associated protein 2), MEC (Muse-Enriched Cell), MSCs (Mesenchymal Stem Cells), Muse (Multilineage-Differentiating Stress-Enduring), NHEJ (Non-Homologous End Joining), ORO (Oil Red O), PBS (Phosphate-Buffered Saline), RT-qPCR (Reverse Transcription Quantitative Polymerase Chain Reaction), SCT (Stress Condition Treatment), SD (Standard Deviation), and SSEA-3 (Stage-Specific Embryonic Antigen-3)
Acknowledgments
We extend our sincere gratitude to provided by the Cellbank of the Stem Cell Institute, University of Science, VNU-HCM, for generously providing the hUCMSCs used in this study.
Author’s contributions
Thuan Minh Le conducted the experiments, acquired the data, analyzed the results, and drafted the manuscript. Ngoc Bao Phan performed experiments and data analysis. Khoi Tuan Truong conducted experiments for cell differentiation. Ngoc Bich Vu critically revised the manuscript for important intellectual content and provided final approval of the version to be published. All authors have read and approved the final manuscript.
Funding
This research is funded by Vietnam National University, Ho Chi Minh City (VNU-HCM) under grant number C2023-18-19.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval
Umbilical cord samples were originally collected under Contract No. 13/HĐ-BVQ2, established between our institution and Le Van Thinh Hospital for a previously approved research project. The collection procedure was reviewed and approved by the hospital’s Ethics Committee, and informed consent was obtained from all donors, including permission for the use of the samples in future research. In the current study, we used human umbilical cord-derived mesenchymal stem cells (hUCMSCs) that had been previously isolated and cryopreserved from these ethically collected samples. All samples were fully anonymized, and no personal identifying information was accessible to the research team. According to institutional policy, further ethical approval was not required for the secondary use of these de-identified samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Velasco
M.G.,
Satué
K.,
Chicharro
D.,
Martins
E.,
Torres-Torrillas
M.,
Peláez
P.,
Multilineage-Differentiating Stress-Enduring Cells (Muse Cells): The Future of Human and Veterinary Regenerative Medicine. Biomedicines.
2023;
11
(2)
:
636
.
View Article PubMed Google Scholar -
Li
J.,
Wu
Z.,
Zhao
L.,
Liu
Y.,
Su
Y.,
Gong
X.,
The heterogeneity of mesenchymal stem cells: an important issue to be addressed in cell therapy. Stem Cell Research & Therapy.
2023;
14
(1)
:
381
.
View Article PubMed Google Scholar -
Mali\vcev
E.,
Jazbec
K.,
An overview of mesenchymal stem cell heterogeneity and concentration. Pharmaceuticals (Basel, Switzerland).
2024;
17
(3)
:
350
.
View Article PubMed Google Scholar -
Kuroda
Y.,
Kitada
M.,
Wakao
S.,
Nishikawa
K.,
Tanimura
Y.,
Makinoshima
H.,
Unique multipotent cells in adult human mesenchymal cell populations. Proceedings of the National Academy of Sciences of the United States of America.
2010;
107
(19)
:
8639-43
.
View Article PubMed Google Scholar -
Wakao
S.,
Kushida
Y.,
Dezawa
M.,
Basic Characteristics of Muse Cells. In: Dezawa, M. (eds) Muse Cells. Advances in Experimental Medicine and Biology, vol 1103. Springer, Tokyo.
2018
.
View Article Google Scholar -
Ogawa
E.,
Oguma
Y.,
Kushida
Y.,
Wakao
S.,
Okawa
K.,
Dezawa
M.,
Naive pluripotent-like characteristics of non-tumorigenic Muse cells isolated from human amniotic membrane. Scientific Reports.
2022;
12
(1)
:
17222
.
View Article PubMed Google Scholar -
Cao
J.,
Yang
Z.,
Xiao
R.,
Pan
B.,
Regenerative potential of pluripotent nontumorgenetic stem cells: multilineage differentiating stress enduring cells (Muse cells). Regenerative Therapy.
2020;
15
:
92-6
.
View Article PubMed Google Scholar -
Yamada
Y.,
Wakao
S.,
Kushida
Y.,
Minatoguchi
S.,
Mikami
A.,
Higashi
K.,
S1P\ S1PR2 axis mediates homing of muse cells into damaged heart for long-lasting tissue repair and functional recovery after acute myocardial infarction. Circulation Research.
2018;
122
(8)
:
1069-83
.
View Article PubMed Google Scholar -
Aprile
D.,
Alessio
N.,
Demirsoy
I.H.,
Squillaro
T.,
Peluso
G.,
Di Bernardo
G.,
MUSE stem cells can be isolated from stromal compartment of mouse bone marrow, adipose tissue, and ear connective tissue: a comparative study of their in vitro properties. Cells.
2021;
10
(4)
:
761
.
View Article PubMed Google Scholar -
Sun
D.,
Yang
L.,
Cao
H.,
Shen
Z.Y.,
Song
H.L.,
Study of the protective effect on damaged intestinal epithelial cells of rat multilineage-differentiating stress-enduring (Muse) cells. Cell Biology International.
2020;
44
(2)
:
549-59
.
View Article PubMed Google Scholar -
Iseki
M.,
Mizuma
M.,
Wakao
S.,
Kushida
Y.,
Kudo
K.,
Fukase
M.,
The evaluation of the safety and efficacy of intravenously administered allogeneic multilineage-differentiating stress-enduring cells in a swine hepatectomy model. Surgery Today.
2021;
51
(4)
:
634-50
.
View Article PubMed Google Scholar -
Castillo
M.G.,
Peralta
T.M.,
Locatelli
P.,
Velazquez
C.,
Herrero
Y.,
Crottogini
A.J.,
Promoting early neovascularization by allotransplanted adipose-derived Muse cells in an ovine model of acute myocardial infarction. PLoS One.
2023;
18
(1)
:
e0277442
.
View Article PubMed Google Scholar -
Yang
Z.,
Liu
J.,
Liu
H.,
Qiu
M.,
Liu
Q.,
Zheng
L.,
Isolation and characterization of SSEA3+ stem cells derived from goat skin fibroblasts. Cellular Reprogramming.
2013;
15
(3)
:
195-205
.
-
Mitani
K.,
Ito
Y.,
Takene
Y.,
Hatoya
S.,
Sugiura
K.,
Inaba
T.,
Long-term trypsin treatment promotes stem cell potency of canine adipose-derived mesenchymal stem cells. Stem Cells and Development.
2021;
30
(6)
:
337-49
.
View Article PubMed Google Scholar -
Dezawa
M.,
Muse cells provide the pluripotency of mesenchymal stem cells: direct contribution of muse cells to tissue regeneration. Cell Transplantation.
2016;
25
(5)
:
849-61
.
View Article PubMed Google Scholar -
Sato
T.,
Wakao
S.,
Kushida
Y.,
Tatsumi
K.,
Kitada
M.,
Abe
T.,
A novel type of stem cells double-positive for SSEA-3 and CD45 in human peripheral blood. Cell Transplantation.
2020;
29
:
963689720923574
.
View Article PubMed Google Scholar -
Heneidi
S.,
Simerman
A.A.,
Keller
E.,
Singh
P.,
Li
X.,
Dumesic
D.A.,
Awakened by cellular stress: isolation and characterization of a novel population of pluripotent stem cells derived from human adipose tissue. PLoS One.
2013;
8
(6)
:
e64752
.
View Article PubMed Google Scholar -
Gimeno
M.L.,
Fuertes
F.,
Barcala Tabarrozzi
A.E.,
Attorressi
A.I.,
Cucchiani
R.,
Corrales
L.,
Pluripotent nontumorigenic adipose tissue-derived muse cells have immunomodulatory capacity mediated by transforming growth factor-β1. Stem Cells Translational Medicine.
2017;
6
(1)
:
161-73
.
View Article PubMed Google Scholar -
Livak
K.J.,
Schmittgen
T.D.,
Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods.
2001;
25
(4)
:
402-8
.
-
Jeter
C.R.,
Liu
B.,
Liu
X.,
Chen
X.,
Liu
C.,
Calhoun-Davis
T.,
NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene.
2011;
30
(36)
:
3833-45
.
View Article PubMed Google Scholar -
Nitobe
Y.,
Nagaoki
T.,
Kumagai
G.,
Sasaki
A.,
Liu
X.,
Fujita
T.,
Neurotrophic factor secretion and neural differentiation potential of multilineage-differentiating stressenduring (Muse) cells derived from mouse adipose tissue. Cell Transplantation.
2019;
28
(9-10)
:
1132-9
.
View Article PubMed Google Scholar -
Kinoshita
K.,
Kuno
S.,
Ishimine
H.,
Aoi
N.,
Mineda
K.,
Kato
H.,
Therapeutic potential of adipose-derived SSEA-3-positive muse cells for treating diabetic skin ulcers. Stem Cells Translational Medicine.
2015;
4
(2)
:
146-55
.
View Article PubMed Google Scholar -
Uchida
H.,
Niizuma
K.,
Kushida
Y.,
Wakao
S.,
Tominaga
T.,
Borlongan
C.V.,
Human muse cells reconstruct neuronal circuitry in subacute lacunar stroke model. Stroke.
2017;
48
(2)
:
428-35
.
View Article PubMed Google Scholar -
Ning
J.,
Cao
Y.Y.,
Zhang
R.Z.,
Li
Y.,
Characteristics of multilineage-differentiating stress-enduring cell clusters in different culture conditions. Skin Research and Technology : Official Journal of International Society for Bioengineering and the Skin (ISBS) [and] International Society for Digital Imaging of Skin (ISDIS) [and] International Society for Skin Imaging (ISSI).
2023;
29
(11)
:
e13528
.
View Article PubMed Google Scholar -
Leng
Z.,
Sun
D.,
Huang
Z.,
Tadmori
I.,
Chiang
N.,
Kethidi
N.,
Quantitative analysis of SSEA3+ cells from human umbilical cord after magnetic sorting. Cell Transplantation.
2019;
28
(7)
:
907-23
.
-
Yamauchi
T.,
Yamasaki
K.,
Tsuchiyama
K.,
Koike
S.,
Aiba
S.,
A quantitative analysis of multilineage-differentiating stress-enduring (Muse) cells in human adipose tissue and efficacy of melanocytes induction. Journal of Dermatological Science.
2017;
86
(3)
:
198-205
.
View Article PubMed Google Scholar -
Aprile
D.,
Alessio
N.,
Squillaro
T.,
Bernardo
G. Di,
Peluso
G.,
Galderisi
U.,
Role of glycosphingolipid SSEA-3 and FGF2 in the stemness and lineage commitment of multilineage differentiating stress enduring (MUSE) cells. Cell Proliferation.
2023;
56
(1)
:
e13345
.
View Article PubMed Google Scholar -
Iseki
M.,
Kushida
Y.,
Wakao
S.,
Akimoto
T.,
Mizuma
M.,
Motoi
F.,
Human muse cells, nontumorigenic phiripotent-like stem cells, have liver regeneration capacity through specific homing and cell replacement in a mouse model of liver fibrosis. Cell Transplantation.
2017;
26
(5)
:
821-40
.
View Article PubMed Google Scholar -
Jögi
A.,
\Ora
I.,
Nilsson
H.,
Lindeheim
A.,
Makino
Y.,
Poellinger
L.,
Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proceedings of the National Academy of Sciences of the United States of America.
2002;
99
(10)
:
7021-6
.
View Article PubMed Google Scholar -
Schmittgen
T.D.,
Zakrajsek
B.A.,
Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. Journal of Biochemical and Biophysical Methods.
2000;
46
(1-2)
:
69-81
.
View Article PubMed Google Scholar -
Zhong
H.,
Simons
J.W.,
Direct comparison of GAPDH, β-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochemical and Biophysical Research Communications.
1999;
259
(3)
:
523-6
.
View Article PubMed Google Scholar -
Alessio
N.,
Squillaro
T.,
Özcan
S.,
Di Bernardo
G.,
Venditti
M.,
Melone
M.,
Stress and stem cells: adult Muse cells tolerate extensive genotoxic stimuli better than mesenchymal stromal cells. Oncotarget.
2018;
9
(27)
:
19328-41
.
View Article PubMed Google Scholar -
Squillaro
T.,
Alessio
N.,
Bernardo
G. Di,
Özcan
S.,
Peluso
G.,
Galderisi
U.,
Stem cells and DNA repair capacity: muse stem cells are among the best performers. Muse Cells: Endogenous Reparative Pluripotent Stem Cells. 2018:103-13.
2018
.
View Article Google Scholar -
Wakao
S.,
Kuroda
Y.,
Ogura
F.,
Shigemoto
T.,
Dezawa
M.,
Regenerative effects of mesenchymal stem cells: contribution of muse cells, a novel pluripotent stem cell type that resides in mesenchymal cells. Cells.
2012;
1
(4)
:
1045-60
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 12 No 4 (2025)
Page No.: 7265-7276
Published on: 2025-04-30
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 612 times
- PDF downloaded - 167 times
- XML downloaded - 37 times
 Biomedpress
Biomedpress