Abstract
Introduction: Breast cancer remains a significant challenge due to disease recurrence and metastasis, often attributed to the persistence of treatment-resistant breast cancer stem cells (BCSCs). Citral, a compound derived from lemongrass essential oil, has demonstrated cytotoxic effects on various cancers, including breast cancer. This study investigates the anticancer mechanisms of Citral in BCSC-enriched 3D cultured spheroids and evaluates its therapeutic potential for estrogen receptor-positive breast cancer.
Methods: Using flow cytometry, the CD44+CD24- population was analyzed, and real-time PCR was employed to measure the expression of ALDH isoforms and pluripotency genes. Additionally, the Nanostring nCounter® PanCancer Pathway Panel was utilized to identify gene expression changes in cancer-related pathways.
Results: Citral treatment significantly reduced spheroid size and the CD44+CD24- stem-like cell population, accompanied by downregulation of ALDH isoforms and pluripotency genes. Gene expression analysis revealed Citral's modulation of key pathways, including PI3K/Akt signaling, cell cycle control, DNA damage response, and apoptosis.
Conclusion: These findings underscore Citral's potential as a promising anticancer agent, particularly for targeting estrogen receptor-positive breast cancer cells and BCSCs. Further preclinical and clinical studies are warranted to explore its therapeutic applications.
Introduction
Worldwide, cancer continues to be the leading cause of death, with almost 20 million new cases and 9.7 million cancer deaths in 20221. The Global Cancer Observatory 2022 (GLOBOCAN 2024) reported that female breast cancer was ranked second with 2.3 million new cases (11.6%), followed by other cancer types. These data indicate that breast cancer is responsible for one out of every four cancer cases and one out of every six cancer deaths1. Despite advancements in treating breast cancer, significant challenges remain, particularly when the disease is detected at advanced stages. Furthermore, disease recurrence and metastasis limit treatment options, leaving patients with an unfavorable prognosis and often resulting in succumbing to the disease. Recurrence is especially challenging in the treatment of estrogen receptor-positive breast cancer, as patients can remain at risk of developing secondary cancer many years after treatment2. The ultimate cause of breast cancer recurrence and metastasis remains unknown3; however, research has identified the presence of breast cancer stem cells (BCSCs) within tumors as the primary drivers4, 5. BCSCs are cells that can regenerate and differentiate, similar to normal stem cells, but are highly tumorigenic6. First identified by Al-Hajj et al.7, this unique cell population expresses surface markers, including CD44+CD24-, aldehyde dehydrogenase (ALDH), and epithelial-specific antigen (ESA) or CD326 (EpCAM)8, 9. The presence of BCSCs in breast cancer hinders treatment, as these cells are reported to be intrinsically resistant to chemotherapy and radiotherapy10, 11. Consequently, non-targeted cancer therapies can eliminate the proliferative and more differentiated cells that comprise the tumor mass. However, treatment-resistant BCSCs possess the capacity to regenerate the tumor gradually, which can lead to cancer relapse due to their self-renewal capability. Therefore, it is crucial to identify alternative approaches to cancer therapy that can directly target BCSCs to prevent cancer recurrence. The use of natural products for cancer prevention and treatment is gaining interest in drug discovery, particularly because they offer a rich source of novel therapeutic agents with minimal side effects12. Current therapy regimens primarily eliminate fast-growing cancer cells, leaving behind quiescent, drug-resistant BCSCs that can subsequently regrow tumors13. Thus, it is essential to identify potential drug candidates that can directly target BCSCs, thereby improving patient survival. Citral (3,7-dimethyl-2,6-octadienal), a naturally occurring essential oil derived from herbal plants such as lemongrass, has been reported to exhibit anticancer effects14, 15, 16. However, its effect on BCSCs remains unclear. Our previous studies have shown that citral can target drug-resistant breast cancer cells17, including its ability to inhibit the expression of the cancer stem cell marker ALDH1A318. Nevertheless, the mechanism behind these effects is still poorly understood, particularly in estrogen receptor-positive breast cancer cells. The organization of cells in a 3D manner provides better insight into the tumorigenesis mechanism of in vitro cancer models, although fully replicating the original tumor microenvironment remains a challenge. The 3D culture system also offers a promising link between traditional 2D cultures and animal models, making it a crucial tool for advancing research in tumor biology19. Therefore, this study explored the anticancer effect of citral on BCSCs and demonstrated that citral is a potent anticancer agent capable of reducing the size of generated breast cancer spheroids using a three-dimensional (3D) culture model. This model is believed to be an effective method for enriching BCSCs from breast cancer cell lines, making it a valuable tool for studying cancer stem cells. In the present work, we discovered that citral inhibits the expression of pluripotency genes and ALDH isoforms and reduces the number of CD44+CD24- stem-like cells. Citral’s mechanism of action on breast cancer spheroids was elucidated through the regulation of various pathways involved in breast cancer, including cell cycle, DNA damage, apoptosis, and stem cell pathways.
Methods
Analysis of Citral by Gas Chromatography-Mass Spectrometry (GCMS)
To determine the purity and components of citral, also known as 3,7-dimethyl-2,6-octadienal (obtained from Sigma Aldrich; CAS 5392-40-5), a study was conducted using a Shimadzu GCMS QP2010 Ultra gas chromatography-mass spectrometry system from Shimadzu Corporation, Japan. The analysis utilized an Rxi™-5ms fused silica capillary column with polarity (30.0 m × 0.25 mm I.D. × 0.25 µm film thickness) from Restek Corporation, Bellefonte, PA, USA. The chromatographic conditions involved a temperature range from 50.0 °C to 300.0 °C (held for 10 minutes) at a rate of 3 °C/min. The sample injection took place at 250 °C using helium (99.9%) as the carrier gas at a flow rate of 0.8 mL/min, with an injection volume of 1 µL and a split ratio of 10:1. In terms of mass spectrometry operation, the ion source was set at a temperature of 200 °C; electron ionization was performed at 70 eV using a mode covering a mass scan range of m/z 40–700. Detection of the components in citral was based on comparing the obtained mass spectra with libraries such as NIST11.lib, NIST11s.lib, FFNSC1.3.lib, and WILEY229.LIB, along with cross-references to existing literature sources. Each component was tentatively quantified based on the relative peak areas in the chromatogram.
Cell Culture
The MCF7 breast cancer cell line (human estrogen receptor-positive), sourced from the human cell culture collection of the UPM-MAKNA Cancer Research Laboratory, Institute of Bioscience, Universiti Putra Malaysia, Malaysia, was used in this study. The cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 incubator (Thermo Fisher Scientific, USA).
Generation of Breast Cancer Spheroids
A 0.6% agarose solution was used to coat cell culture vessels after solidification for 30 minutes at ambient temperature under sterile conditions. To generate breast cancer spheroids, breast cancer cells were cultured in the agarose-coated wells in serum-free DMEM/F-12 medium containing insulin (10 µg/mL), gentamycin (10 µg/mL), human fibroblast growth factor (hFGF) (0.1 µg/mL), hydrocortisone (1 µg/mL), antibiotic-antimycotic (50×), and B27 supplement (50×). Spheroids were grown for 5 days in a CO₂ incubator without disturbing the culture plates and analyzed using an inverted microscope.
CellTiter-Glo® 3D Assay
The cytotoxicity of citral on breast cancer spheroids was assayed using the Promega CellTiter-Glo® 3D reagent (Promega, USA). The CellTiter-Glo® 3D assay is an ATP-based assay that quantifies luminescent signals produced by the cells. This assay incorporates lytic activity to enhance penetration into cell aggregates such as spheroids, allowing full quantification of cell viability20. In each well of a 96-well plate coated with agarose, 3,000 cells were plated in 100 µL of spheroid growth medium. Breast cancer spheroids were formed by culturing the cells for 48 hours. These spheroids were then treated with concentrations of citral ranging from 0 µM to 500 µM for 72 hours. After treatment, CellTiter-Glo® 3D solution was added to lyse the spheroids by shaking them for 5 minutes in a microplate reader. The plate was incubated at room temperature for 25 minutes before measuring the luminescence signal using a microplate reader. IC₅₀ values for citral on breast cancer spheroids were determined by creating a dose-response curve using GraphPad Prism 7.0 software. The identified IC₅₀ was used for subsequent experiments, and the effects of citral treatment were compared to the untreated group. The IC₅₀ is commonly used to measure the drug concentration required to inhibit 50% of a biological process, helping researchers understand the drug's potency in pharmacological research21. The CellTiter-Glo® 3D experiments were conducted three times (n=3).
Effects of Breast Cancer Spheroids after Citral Treatment
Following the 72-hour citral treatment, breast cancer spheroids of MCF7 were observed under an inverted microscope. ZEISS ZenPro software was used to capture images of breast cancer spheroids that were both untreated and treated with citral. Subsequently, the size of the spheroids was measured using ImageJ software22. The size measurements obtained from both spheroids were recorded, and the results were analyzed by an unpaired t-test using GraphPad Prism 7.0 software.
Immunophenotyping of CD44+/CD24- Breast Cancer Stem Cells
Two surface markers, CD44+ and CD24-, were further assessed to identify the population of breast cancer stem cells (BCSCs). The staining steps for immunophenotyping of CD44+CD24- BCSCs were conducted by incubating the cell suspension with conjugated antibodies on ice and protected from light for 30 minutes. Following the manufacturer’s recommendations, the conjugated antibodies were diluted in FACS buffer to prepare antibody solutions. The cell suspension was stained concurrently with both antibodies for double staining (CD44-FITC/CD24-PE). Following incubation, the cells were collected by centrifugation at 1000 rpm for 5 minutes. After discarding the supernatant, the cells were washed once with ice-cold FACS buffer. To ensure a uniform, single-cell population, the cells were resuspended in 1 mL of FACS buffer and passed through a 40 µm cell strainer into FACS tubes. Stained cells were kept on ice and in the dark before being analyzed. A BD Fortessa® flow cytometer was utilized to assess the percentage of stained cells.
| No | Gene | NCBI Gene ID | Name* | 5'---3' Sequence | Length | GC % | Tm | Size (bp) |
| 1 | ALDH1A1 | NM_000689.5 | f_ALDH1A1 | TGGACCAGTGCAGCAAATCA | 20 | 50 | 60.18 | 171 |
| r_ALDH1A1 | ACGCCATAGCAATTCACCCA | 20 | 50 | 60.03 | ||||
| 2 | ALDH1A3 | NM_000693.4 | f_ALDH1A3 | TGGCACGAATCCAAGAGTGG | 20 | 55 | 60.32 | 101 |
| r_ALDH1A3 | TTGTCCACGTCGGGCTTATC | 20 | 55 | 60.11 | ||||
| 3 | ALDH2 | NM_000690.4 | f_ALDH2 | TTCTTCAACCAGGGCCAGTG | 20 | 55 | 60.18 | 200 |
| r_ALDH2 | TTCCCCGTGTTGATGTAGCC | 20 | 55 | 60.04 | ||||
| 4 | NANOG | NM_024865.4 | f_NANOG | AATACCTCAGCCTCCAGCAGATG | 23 | 52.17 | 62.08 | 148 |
| r_NANOG | TGCGTCACACCATTGCTATTCTTC | 24 | 45.83 | 61.92 | ||||
| 5 | MYC | NM_002467.6 | f_MYC | CATCAGCACAACTACGCAGC | 20 | 55 | 59.9 | 120 |
| r_MYC | GCTGGTGCATTTTCGGTTGT | 20 | 50 | 59.97 | ||||
| 6 | SOX2 | NM_003106.4 | f_SOX2 | GCCCTGCAGTACAACTCCAT | 20 | 55 | 60.04 | 128 |
| r_SOX2 | GACTTGACCACCGAACCCAT | 20 | 55 | 59.96 | ||||
| 7 | OCT3/4 | NM_002701.6 | f_OCT3/4 | CAAAGCAGAAACCCTCGTGC | 20 | 55 | 60.04 | 171 |
| r_OCT3/4 | AACCACACTCGGACCACATC | 20 | 55 | 59.97 | ||||
| 8 | GAPDH** | NM_002046.7 | f_GAPDH | GTCATCCCTGAGCTGAACGG | 20 | 60 | 60.46 | 187 |
| r_GAPDH | CCACCTGGTGCTCAGTGTAG | 20 | 60 | 60.04 |
Quantitative Reverse Transcriptase Polymerase Chain Reaction (RT-qPCR)
All primers were designed using the online NCBI Primer-Blast tool available at https://www.ncbi.nlm.nih.gov/tools/primer-blast/ and were referenced to the NCBI Gene database. The specificity of PCR amplification was ensured based on guidelines for RT-qPCR primer design provided by Integrated DNA Technologies (IDT), Singapore, available online. All primers (Table 1) were synthesized by Integrated DNA Technologies (IDT), Singapore. The melting temperature (Tm) of each primer pair was optimized by conventional PCR. Total RNA from breast cancer spheroid cells was isolated using the Total RNA Isolation Kit (Pure Direx, Bio-Helix, Taiwan), and cDNA was prepared using the Tetro™ cDNA Synthesis Kit (Bioline, UK). The synthesized cDNA was prepared before RT-qPCR experiments or stored at -20 °C for long-term preservation.
The qPCR was conducted using a Bio-Rad CFX96 (Bio-Rad, USA) thermocycler, and the analysis was performed using Bio-Rad CFX Manager software version 3.1. The reaction mix was prepared using the SensiFAST™ SYBR® No ROX Kit (Bioline) as recommended by the manufacturer’s protocols. The RT-qPCR reaction volume for each sample was 20 µL in 0.2 mL PCR tubes. All reactions were conducted in triplicate (n = 3). All data acquired were analyzed for quantification, melt curve, and gene expression. RT-qPCR was repeated for three independent experiments, and GraphPad Prism 7.0 and Microsoft Excel were utilized for data analysis.
Multiplex Gene Pathway Analysis
The Nanostring nCounter® PanCancer Pathway Panel is a multiplex gene analysis platform that analyzes 770 genes from 13 cancer-associated canonical pathways. In this study, the pathway panel was used to measure the treatment effects of citral on the regulation of cancer pathways in breast cancer spheroids. The nCounter® PanCancer Pathways Panel (Nanostring, USA) was utilized to assess the effects of citral on breast cancer spheroids (n = 3 for each group: untreated and citral-treated MCF7 spheroids) related to cancer-associated canonical pathways. All procedures were conducted following the manufacturer’s instructions. The procedure started with hybridizing the Nanostring™ probe with 50 ng of total RNA extracted from each sample at 65 °C overnight (16–24 hours).
Following that, hybridized samples were analyzed, and readout data obtained were processed using the manufacturer’s software, nSolver® Analysis Software, to check quality control (QC), mRNA positive control QC, and normalization QC. Genes with >2-fold change in their expression, according to Benjamini & Yekutieli23, with a p-value < 0.05 were considered significant. Differential expression and pathway scoring were performed using a built-in nSolver® advanced analysis module. Gene Ontology and pathway analyses were conducted using the software WebGestalt (https://www.webgestalt.org).
Statistical Analysis
The experiments were performed in triplicate, and the results were expressed as mean ± standard deviation (SD). Data analysis utilized Microsoft Office Excel 2019 and GraphPad Prism 7.0. Statistical significance was defined as a p-value less than 0.05 (p < 0.05).
| Peak | Compound | Library | LRI | SI (%) | Composition (%) | Reference |
| 1 | trans -Rose oxide | FFNSC1.3.lib | 1133 | 83 | 0.60 | Babushok, et al. 2011 24 |
| 2 | cis -Isocitral | FFNSC1.3.lib | 1164 | 92 | 0.80 | Adams, 2007 25 |
| 3 | trans -Isocitral | FFNSC1.3.lib | 1182 | 93 | 1.37 | Adams, 2007 25 |
| 4 | Neral * | FFNSC1.3.lib | 1247 | 94 | 43.43 | Babushok et al ., 2011 24 |
| 5 | Geranial ** | WILEY229.lib | 1277 | 95 | 44.35 | Babushok et al ., 2011 24 |
| 6 | UNKNOWN | N/A*** | 1285 | N/A | 1.30 | N/A |
| 7 | Neryl dimethyl acetal | WILEY229.lib | 1318 | 83 | 3.25 | Yannai, 2012 26 |
| 8 | Geranyl dimethyl acetal | WILEY229.lib | 1342 | 87 | 4.90 | Yannai, 2012 26 |
Results
Evaluation of Citral by GC-MS
The purity and composition of citral (C10H16O) were first assessed by mass spectral GC-MS analysis through the detection of the retention time of each peak. The identification was performed by comparing the compounds' calculated linear retention indices (LRI) to the reported LRI values in the literature (Table 2 ). The mass spectra from the GC-MS analysis were matched against the reference spectra in Wiley’s Flavors and Fragrances of Natural and Synthetic Compounds (FFNSC) 1.3 and WILEY229 GC-MS libraries. The GC-MS analysis revealed two major constituents of citral: neral (cis-citral) and geranial (trans-citral) (Figure 1), with percentage compositions of 43.43% and 44.35%, respectively, and an overall purity of 87.7%. Hence, it confirms the suitability of citral for treating breast cancer spheroids.
Effect of Citral on the Proliferation of Breast Cancer Spheroids
After assessing the purity of citral, breast cancer spheroids were exposed to increasing concentrations of the compound, and the viability of cells was assessed after 72 hours. This identified an IC50 (concentration of compound required for 50% inhibition of cell viability) value of 61.09 µM ± 6.8. Treatment with citral at this concentration was able to reduce the proliferation of breast cancer spheroids, with a significant size reduction (Figure 2).
Inhibition of Cancer Stemness in Breast Cancer Spheroids Following Citral Treatment
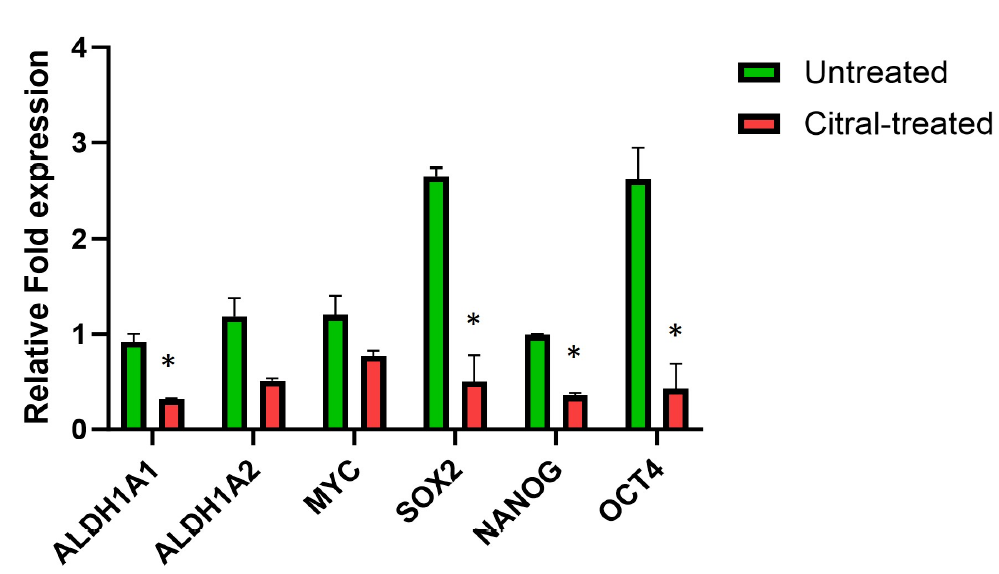
To study the effect of citral on the BCSC population, the expression of CD44+CD24- cells was next analyzed. As shown in Figure 3, citral reduced the number of CD44+CD24- breast cancer cells following 72 hours of treatment. The percentage of CD44+CD24- breast cancer cells in MCF7 spheroids was significantly decreased from 13.2 ± 0.75% to 5.4 ± 0.45% (p < 0.05). Furthermore, no statistical difference was measured within the CD44+CD24+ or CD44-CD24+ populations, suggesting the effect of citral may be due to defective proliferation rather than induction of cancer stem cell differentiation. We next evaluated the regulation of stemness genes in the breast cancer spheroids. Treatment with citral significantly downregulated the expression of ALDH1 and the pluripotency genes SOX2, NANOG, and OCT4 (Figure 4).
Analysis of Gene Expression of Canonical Pathways Regulated by Citral
Given the major effect of citral on MCF-7 breast cancer spheroids, we next evaluated the expression of cancer pathways in this cell line using the Nanostring nCounter® PanCancer Pathway Panel. Two-way hierarchical clustering was conducted on both mRNA (rows) and biological replicates (columns) as a quality control measure to identify outliers, as it groups samples based on the similarity of their gene expression patterns27.
A total of six samples (3 untreated samples and 3 citral-treated samples) were used for each breast cancer spheroid, and replicates were consistently clustered together between the untreated and citral-treated groups. Treatment with citral resulted in a change in the expression of 52 genes, with 46 genes being downregulated and 6 genes upregulated (Figure 5).
Gene ontology analysis highlighted negative regulation of transferase activity, epithelial cell proliferation, and cell homeostasis as the most significant biological processes affected by citral. In addition, the most significant pathways were pathways in cancer, PI3K/Akt signaling, DNA damage, cell cycle, and apoptosis signaling pathways (Figure 5). We found a significant downregulation of genes implicated in controlling the cell cycle and proliferation of epithelial cells. The inhibitor of apoptosis, BCL-2, was also downregulated. Importantly, treatment with citral induced downregulation of several additional genes involved in cancer stemness, including LIFR, STAT3, SUV39H2, IBSP, and ID4 (Figure 6).
Discussion
For cancer treatment, the presence of cancer stem cells (CSCs) poses a significant therapeutic obstacle. Conventional chemotherapy causes tumor shrinkage at the primary tumor site and simultaneously triggers stress responses to protect a specific subset of cells against the drug activity. Such stimuli induce the generation of chemo-resistant cells that closely align with the phenotype and properties of CSCs28, 29. Therefore, there is a critical need to find new therapies targeting these resistant cells, which are responsible for cancer recurrence. The preclinical model of 3D spheroid culture is an effective method to enrich for breast cancer stem cells (BCSCs) and derived progenitors, as only CSCs can proliferate in anchorage-independent conditions and the absence of serum in the medium30. Under these conditions, the unique subpopulation of cells enriched in spheroid culture with stem-like properties can self-renew and are highly tumorigenic. In this study, we have utilized the MCF-7 cell line as an in vitro model for estrogen receptor-positive breast cancer to test the effectiveness of the naturally abundant compound citral in inhibiting BCSC proliferation as a strategy for new treatments of breast cancer with a high prevalence of recurrence. Ponti et al.31 demonstrated that spheroid culture using the MCF-7 cell line enriched a population of cells with a protein expression signature similar to stem cells and was capable of forming tumors from as few as 1,000 cells when injected into the mammary fat pad of mice.
Citral was found to exert an anticancer effect by reducing spheroid size and significantly decreasing the number of CD44+CD24- breast cancer stem cells in MCF-7 spheroids. Additionally, it was observed that the expression of several pluripotency genes—SOX2, OCT4, NANOG, and ALDH1 isoforms—was reduced after citral treatment. OCT4, NANOG, and SOX2 are transcription factors that are expressed in both CSCs and embryonic stem cells (ESCs) and are related to pluripotency and stemness32. The overexpression of these three transcription factors in cancer cells induces tumorigenicity, tumor metastasis, and recurrence after chemotherapy33.
Furthermore, overexpression of aldehyde dehydrogenase (ALDH) in breast cancer cells has been shown to demonstrate stem/progenitor properties8. Therefore, targeting specific ALDH isoforms may help to eliminate the CSC subpopulation, thereby reducing the likelihood of cancer recurrence. Research conducted by Coyle et al.34 demonstrated that an isoform of the ALDH1 family (ALDH1A1) significantly regulates the proliferation and differentiation of cancer cells via the oxidation of retinol to retinoic acid (RA). Surprisingly, high ALDH1 expression in breast cancer is correlated with a poor prognosis35.
Therefore, the ability of citral to reduce the expression of pluripotency genes and ALDH1 genes, including the population of tumor-initiating CD44+CD24- cells, indicates its potential in specifically targeting the BCSC subpopulation within tumor spheroids.
Additionally, this study aimed to elucidate the mechanism responsible for citral’s effect on breast cancer spheroids. Several genes in pathways related to cell cycle regulation and cell growth were significantly downregulated in the spheroids treated with citral, suggesting an effect mediated by cell cycle arrest and reduction of epithelial cell proliferation. Important components involved in cell cycle progression are the cell cycle cyclin-kinase complexes CDK4/6-cyclin D and the transcription factor E2F5. Cyclin-dependent kinases (CDKs), such as CDK4 and CDK6, are well recognized to govern cell cycle activity and are also strongly linked to the development and progression of tumors36. Treatment with citral downregulated CDK6, CCNE1, and E2F5 genes, thus limiting cell proliferation. The formation of cyclin-CDK complexes facilitates control of cell cycle progression by phosphorylating targets such as RB1, which was also found to be downregulated37.
The expression of GSK3B was instead upregulated. GSK3 limits cell cycle progression by phosphorylation of cyclin D1, thus tagging the protein for degradation. DUSP5, which can negatively regulate members of the proliferation pathways activated by the mitogen-activated protein (MAP) kinase superfamily (MAPK/ERK, SAPK/JNK, p38), was also upregulated and functions as a tumor suppressor38, 39. In addition, the key inhibitor of apoptosis, BCL2, was found to be downregulated. Low expression of BCL2 can trigger caspase enzymes and promote cell apoptosis40.
Together with a marked effect on cell proliferation, citral downregulated other pathways associated with the regulation of cancer stemness and recurrence. The LIFR and STAT3 genes were among the most downregulated genes after treatment with citral. The expression of LIFR and STAT3 is correlated with stem cell dormancy and relapse in estrogen receptor-positive breast cancer. Their loss has been demonstrated to reduce the expression of cancer stem cell-associated genes41. The same level of downregulation was also found for the ID4 gene, which maintains BCSC self-renewal42. Finally, the downregulation of the oncogene SUV39H2 and IBSP has been observed to decrease the metastatic capability of breast cancer cells and, therefore, the incidence of cancer recurrence43, 44.
Despite some study limitations, including that the spheroid model used may not fully replicate the complexity of the tumor microenvironment and cancer stem cell heterogeneity, our data demonstrate that citral may be a good candidate therapeutic compound targeting breast cancer stem cells that can complement other drugs with similar potential. Several BCSC-targeting agents include salinomycin, curcumin, and resveratrol45, 46. Salinomycin inhibits the Wnt/β-catenin signaling pathway, which is crucial for BCSC maintenance and proliferation. Its mechanism involves disrupting the CSC microenvironment and promoting CSC apoptosis47. Meanwhile, curcumin, derived from turmeric, inhibits the proliferation of breast cancer stem cells (BCSCs), suppresses metastasis, and ultimately limits tumor formation48. Finally, resveratrol affects BCSCs via multiple signaling pathways, including suppression of Wnt/β-catenin signaling and induction of autophagy49.
Further studies could investigate the effect of citral in combination with other compounds and the long-term toxicity of citral compared to other agents. While citral is natural and generally considered safe, its full safety profile in cancer therapy remains to be thoroughly investigated.
To further validate the exact mechanism of citral, some suggestions are proposed for future investigation. Several studies have suggested improved methods for enriching breast cancer stem cells in vitro. The current spheroid culture can be improved by creating a hypoxic environment to further enrich the BCSC population and by isolating BCSCs using fluorescence-activated cell sorting. In addition, the introduction of other tumor microenvironment factors to the in vitro culture can be achieved by co-culturing breast cancer stem cells with stromal cells such as fibroblasts and macrophages to better recapitulate the tumor niche. Finally, high-throughput molecular biology techniques such as next-generation sequencing and proteomics could be applied to further investigate the anticancer potential of citral.
Conclusion
In conclusion, our data point to a significant inhibitory effect on cell proliferation and stem cell pathways. Therefore, the ability of citral to limit BCSC proliferation is of great importance for developing new therapeutic strategies for both the treatment and prevention of breast cancer. Citral was found to exert its anticancer mechanism, as determined by the reduction of spheroid size, induction of apoptosis, and a decrease in the percentage of ALDH+ and CD44+CD24- breast cancer stem cell populations. Furthermore, the current study showed that citral was able to inhibit the expression of pluripotency and ALDH isoforms that regulate the stemness property of BCSCs. These in vitro findings provide a rationale for further preclinical and clinical evaluation of citral for anti-BCSC therapy. Although the exact underlying mechanism of citral in targeting BCSCs was not conclusively determined using the Nanostring™ platform, the outcome has provided important insights, especially into cell proliferation and stem cell pathways. However, although the pathways affected by citral require further validation by Western blotting and real-time PCR, the outcomes of this study have shown that citral exhibits compelling potential in targeting BCSCs.
Abbreviations
ALDH - Aldehyde Dehydrogenase, BCL-2 - B-Cell Lymphoma 2 (an anti-apoptotic protein), BCSCs - Breast Cancer Stem Cells, CCNE1 - Cyclin E1, CD44+CD24- - A surface marker phenotype associated with breast cancer stem cells, CDK - Cyclin-Dependent Kinase, cDNA - Complementary DNA, CSCs - Cancer Stem Cells, DMEM/F-12 - Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12, DNA - Deoxyribonucleic Acid, DUSP5 - Dual Specificity Phosphatase 5, E2F5 - E2F Transcription Factor 5, EpCAM - Epithelial Cell Adhesion Molecule (CD326), ESA - Epithelial-Specific Antigen, ESCs - Embryonic Stem Cells, FACS - Fluorescence-Activated Cell Sorting, FBS - Fetal Bovine Serum, GC-MS - Gas Chromatography-Mass Spectrometry, GSK3B - Glycogen Synthase Kinase 3 Beta, hFGF - Human Fibroblast Growth Factor, IBSP - Integrin-Binding Sialoprotein, IC₅₀ - Half Maximal Inhibitory Concentration, ID4 - Inhibitor of DNA Binding 4, LIFR - Leukemia Inhibitory Factor Receptor, MAPK/ERK - Mitogen-Activated Protein Kinase/Extracellular Signal-Regulated Kinase, mRNA - Messenger RNA, NANOG - A key pluripotency transcription factor, nSolver® - Software for analyzing Nanostring data, OCT4 - Octamer-Binding Transcription Factor 4, PI3K/Akt - Phosphoinositide 3-Kinase/Protein Kinase BQC - Quality Control, RA - Retinoic Acid, RB1 - Retinoblastoma 1 (a tumor suppressor gene), RNA - Ribonucleic Acid, RT-qPCR - Reverse Transcription Quantitative Polymerase Chain Reaction, SAPK/JNK - Stress-Activated Protein Kinase/c-Jun N-Terminal Kinase, SOX2 - SRY-Box Transcription Factor 2, STAT3 - Signal Transducer and Activator of Transcription 3, SUV39H2 - Suppressor of Variegation 3-9 Homolog 2, Tm - Melting Temperature, WebGestalt - WEB-based GEne SeT AnaLysis Toolkit, Wnt/β-catenin - A signaling pathway involved in stem cell maintenance and cancer progression
Acknowledgments
The authors thank the management of the UPM MAKNA Cancer Research Laboratory, Institute of Bioscience UPM, for providing access to the facilities during the project.
Author’s contributions
Muhammad Ehsan Fitri Rusli: Conducting experiments & writing. Rozita Rosli: Conceptualization, Writing - Review & Editing, Supervision. Cinzia Allegrucci: Conceptualization, Writing - Review & Editing. Norazalina Saad: Conceptualization, Funding acquisition, Writing-review & editing, Supervision, Project administration. All authors read and approved the final manuscript.
Funding
The study was funded by Universiti Putra Malaysia in Serdang, Selangor, Malaysia (UPM/GP IPM/9570800).
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Bray
F.,
Laversanne
M.,
Sung
H.,
Ferlay
J.,
Siegel
R.L.,
Soerjomataram
I.,
Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians.
2024;
74
(3)
:
229-63
.
View Article PubMed Google Scholar -
Pedersen
R.N.,
Esen
B.Ö.,
Mellemkj\aer
L.,
Christiansen
P.,
Ejlertsen
B.,
Lash
T.L.,
The Incidence of Breast Cancer Recurrence 10-32 Years After Primary Diagnosis. JNCI: Journal of the National Cancer Institute.
2021;
114
(3)
:
391-9
.
View Article Google Scholar -
Fares
J.,
Fares
M.Y.,
Khachfe
H.H.,
Salhab
H.A.,
Fares
Y.,
Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduction and Targeted Therapy.
2020;
5
(1)
:
28
.
View Article PubMed Google Scholar -
Ahmad
A.,
Pathways to breast cancer recurrence. ISRN Oncology.
2013;
2013
:
290568
.
View Article PubMed Google Scholar -
McDermott
S.P.,
Wicha
M.S.,
Targeting breast cancer stem cells. Molecular Oncology.
2010;
4
(5)
:
404-19
.
View Article PubMed Google Scholar -
Beck
B.,
Blanpain
C.,
Unravelling cancer stem cell potential. Nature Reviews. Cancer.
2013;
13
(10)
:
727-38
.
View Article PubMed Google Scholar -
Al-Hajj
M.,
Wicha
M.S.,
Benito-Hernandez
A.,
Morrison
S.J.,
Clarke
M.F.,
Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America.
2003;
100
(7)
:
3983-8
.
View Article PubMed Google Scholar -
Ginestier
C.,
Hur
M.H.,
Charafe-Jauffret
E.,
Monville
F.,
Dutcher
J.,
Brown
M.,
ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell.
2007;
1
(5)
:
555-67
.
View Article PubMed Google Scholar -
Lin
C.Y.,
Barry-Holson
K.Q.,
Allison
K.H.,
Breast cancer stem cells: are we ready to go from bench to bedside?. Histopathology.
2016;
68
(1)
:
119-37
.
View Article PubMed Google Scholar -
Pavlopoulou
A.,
Oktay
Y.,
Vougas
K.,
Louka
M.,
Vorgias
C.E.,
Georgakilas
A.G.,
Determinants of resistance to chemotherapy and ionizing radiation in breast cancer stem cells. Cancer Letters.
2016;
380
(2)
:
485-93
.
View Article PubMed Google Scholar -
Wicha
M.S.,
Liu
S.,
Dontu
G.,
Cancer stem cells: an old idea - a paradigm shift. Cancer Research.
2006;
66
(4)
:
1883-90
.
View Article PubMed Google Scholar -
Reddy
L.,
Odhav
B.,
Bhoola
K.D.,
Natural products for cancer prevention: a global perspective. Pharmacology & Therapeutics.
2003;
99
(1)
:
1-13
.
View Article PubMed Google Scholar -
Lindell
E.,
Zhong
L.,
Zhang
X.,
Quiescent Cancer Cells-A Potential Therapeutic Target to Overcome Tumor Resistance and Relapse. International Journal of Molecular Sciences.
2023;
24
(4)
:
3762
.
View Article PubMed Google Scholar -
Dosoky
N.S.,
Setzer
W.N.,
Biological Activities and Safety of Citrus spp. Essential Oils. International Journal of Molecular Sciences.
2018;
19
(7)
:
1-25
.
View Article PubMed Google Scholar -
Chaouki
W.,
Leger
D.Y.,
Liagre
B.,
Beneytout
J.L.,
Hmamouchi
M.,
Citral inhibits cell proliferation and induces apoptosis and cell cycle arrest in MCF-7 cells. Fundamental & Clinical Pharmacology.
2009;
23
(5)
:
549-56
.
View Article PubMed Google Scholar -
Sanches
L.J.,
Marinello
P.C.,
Panis
C.,
Fagundes
T.R.,
Morgado-Díaz
J.A.,
de-Freitas-Junior
J.C.,
Cytotoxicity of citral against melanoma cells: the involvement of oxidative stress generation and cell growth protein reduction. Tumour Biology.
2017;
39
(3)
:
1010428317695914
.
View Article PubMed Google Scholar -
Nigjeh
S.E.,
Yeap
S.K.,
Nordin
N.,
Kamalideghan
B.,
Ky
H.,
Rosli
R.,
Citral induced apoptosis in MDA-MB-231 spheroid cells. BMC Complementary and Alternative Medicine.
2018;
18
(1)
:
56
.
View Article PubMed Google Scholar -
Thomas
M.L.,
de Antueno
R.,
Coyle
K.M.,
Sultan
M.,
Cruickshank
B.M.,
Giacomantonio
M.A.,
Citral reduces breast tumor growth by inhibiting the cancer stem cell marker ALDH1A3. Molecular Oncology.
2016;
10
(9)
:
1485-96
.
View Article PubMed Google Scholar -
Poornima
K.,
Francis
A.P.,
Hoda
M.,
Eladl
M.A.,
Subramanian
S.,
Veeraraghavan
V.P.,
Implications of Three-Dimensional Cell Culture in Cancer Therapeutic Research. Frontiers in Oncology.
2022;
12
(May)
:
891673
.
View Article PubMed Google Scholar -
Kijanska
M.,
Kelm
J.,
In vitro 3D Spheroids and Microtissues: ATP-based Cell Viability and Toxicity Assays. Assay guidance manual. Bethesda, MD: Eli Lilly & Company.
2004
.
-
Kristi
L.,
Determination of half-maximal inhibitory concentration using biosensor-based protein interaction analysis. Physiology & Behavior.
2017;
176
(3)
:
139-48
.
View Article Google Scholar -
Rueden
C.T.,
Schindelin
J.,
Hiner
M.C.,
DeZonia
B.E.,
Walter
A.E.,
Arena
E.T.,
ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics.
2017;
18
(1)
:
529
.
View Article PubMed Google Scholar -
Benjamini
Y.,
Yekutieli
D.,
The Control of the False Discovery Rate in Multiple Testing under Dependency. 2001;
:
1165-88
.
View Article Google Scholar -
Babushok
V.I.,
Linstrom
P.J.,
Zenkevich
I.G.,
Retention Indices for Frequently Reported Compounds of Plant Essential Oils. J Phys Chem Ref Data.
2011;
40
(4)
:
043101
.
View Article Google Scholar -
Yannai
S.,
Dictionary of Food Compounds with CD-ROM. 2nd ed. CRC Press; 2012. https://doi.org/10.1201/b12964.CRC Press 2012.
View Article Google Scholar -
Adams
R.P.,
Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry (4th ed.). Carol Stream: Allured Publishing Corporation. 2007.
Google Scholar -
Zhao
S.,
Guo
Y.,
Sheng
Q.,
Shyr
Y.,
Advanced heat map and clustering analysis using heatmap3. BioMed research international.
2014;
2014
:
986048
.
View Article PubMed Google Scholar -
Pinto
B.,
Henriques
A.C.,
Silva
P.M.,
Bousbaa
H.,
Three-dimensional spheroids as in vitro preclinical models for cancer research. Pharmaceutics.
2020;
12
(12)
:
1-38
.
View Article PubMed Google Scholar -
De Angelis
M.L.,
Francescangeli
F.,
La Torre
F.,
Zeuner
A.,
Stem cell plasticity and dormancy in the development of cancer therapy resistance. Front Oncol.
2019;
9
(JULY)
:
626
.
View Article PubMed Google Scholar -
Steinbichler
T.B.,
Dudás
J.,
Skvortsov
S.,
Ganswindt
U.,
Riechelmann
H.,
Skvortsova
I.I.,
Therapy resistance mediated by cancer stem cells. Semin Cancer Biol.
2018;
53
:
156-67
.
View Article PubMed Google Scholar -
Ponti
D.,
Costa
A.,
Zaffaroni
N.,
Pratesi
G.,
Petrangolini
G.,
Coradini
D.,
Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res.
2005;
65
(13)
:
5506-11
.
View Article PubMed Google Scholar -
Liu
A.,
Yu
X.,
Liu
S.,
Pluripotency transcription factors and cancer stem cells: small genes make a big difference. Chin J Cancer.
2013;
32
(9)
:
483-7
.
View Article PubMed Google Scholar -
Ben-Porath
I.,
Thomson
M.W.,
Carey
V.J.,
Ge
R.,
Bell
G.W.,
Regev
A.,
An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet.
2008;
40
(5)
:
499-507
.
View Article PubMed Google Scholar -
Coyle
K.M.,
Sultan
M.,
Thomas
M.L.,
Vaghar-Kashani
A.,
Marcato
P.,
Retinoid Signaling in Cancer and Its Promise for Therapy. J Carcinog Mutagen.
2013;
S7
:
1-14
.
View Article Google Scholar -
Xing
P.,
Dong
H.,
Liu
Q.,
Zhao
T.,
Yao
F.,
Xu
Y.,
ALDH1 Expression and Vasculogenic Mimicry Are Positively Associated with Poor Prognosis in Patients with Breast Cancer. Cell Physiol Biochem.
2018;
49
(3)
:
961-70
.
View Article PubMed Google Scholar -
Choi
Y.J.,
Li
X.,
Hydbring
P.,
Sanda
T.,
Stefano
J.,
Christie
A.L.,
The requirement for cyclin D function in tumor maintenance. Cancer Cell.
2012;
22
(4)
:
438-51
.
View Article PubMed Google Scholar -
Ding
L.,
Cao
J.,
Lin
W.,
Chen
H.,
Xiong
X.,
Ao
H.,
The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci.
2020;
21
(6)
:
1-28
.
View Article PubMed Google Scholar -
Cai
C.,
Chen
J.,
Han
Z.,
He
H.,
Chen
J.,
Chen
Y.,
Down-regulation of dual-specificity phosphatase 5 predicts poor prognosis of patients with prostate cancer. International journal of clinical and experimental medicine.
2015;
8
(3)
:
4186-4194
.
PubMed Google Scholar -
de Oliveira
G.A.,
Rangel
L.P.,
Costa
D.C.,
Silva
J.L.,
Misfolding, aggregation, and disordered segments in c-Abl and p53 in human cancer. Front Oncol.
2015;
5
(APR)
:
97
.
View Article PubMed Google Scholar -
Fatemizadeh
M.,
Tafvizi
F.,
Shamsi
F.,
Amiri
S.,
Farajzadeh
A.,
Akbarzadeh
I.,
Apoptosis Induction, Cell Cycle Arrest and Anti-Cancer Potential of Tamoxifen-Curcumin Loaded Niosomes Against MCF-7 Cancer Cells. Iran J Pathol.
2022;
17
(2)
:
183-90
.
View Article PubMed Google Scholar -
Johnson
R.W.,
Finger
E.C.,
Olcina
M.M.,
Vilalta
M.,
Aguilera
T.,
Miao
Y.,
Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nat Cell Biol.
2016;
18
(10)
:
1078-89
.
View Article PubMed Google Scholar -
Junankar
S.,
Baker
L.A.,
Roden
D.L.,
Nair
R.,
Elsworth
B.,
Gallego-Ortega
D.,
ID4 controls mammary stem cells and marks breast cancers with a stem cell-like phenotype. Nat Commun.
2015;
6
(1)
:
6548
.
View Article PubMed Google Scholar -
Li
B.,
Zheng
Y.,
Yang
L.,
The oncogenic potential of SuV39H2: A comprehensive and perspective view. Journal of Cancer.
2019;
10
(3)
:
721-9
.
View Article PubMed Google Scholar -
Wu
K.,
Feng
J.,
Lyu
F.,
Xing
F.,
Sharma
S.,
Liu
Y.,
Exosomal miR-19a and IBSP cooperate to induce osteolytic bone metastasis of estrogen receptor-positive breast cancer. Nature communications.
2021;
12
(1)
:
5196
.
View Article PubMed Google Scholar -
Chu
M.,
Zheng
C.,
Chen
C.,
Song
G.,
Hu
X.,
Wang
Z.W.,
Targeting cancer stem cells by nutraceuticals for cancer therapy. Seminars in cancer biology.
2022;
85
:
234-45
.
View Article PubMed Google Scholar -
Palomeras
S.,
Ruiz-Martínez
S.,
Puig
T.,
Targeting breast cancer stem cells to overcome treatment resistance. Molecules.
2018;
23
(9)
:
2193
.
View Article PubMed Google Scholar -
Wang
H.,
Zhang
H.,
Zhu
Y.,
Wu
Z.,
Cui
C.,
Cai
F.,
Anticancer Mechanisms of Salinomycin in Breast Cancer and Its Clinical Applications. Frontiers in oncology.
2021;
11
(July)
:
654428
.
View Article PubMed Google Scholar -
Liu
H.T.,
Ho
Y.S.,
Anticancer effect of curcumin on breast cancer and stem cells. Food Sci Hum Wellness.
2018;
7
(2)
:
134-7
.
View Article Google Scholar -
Nguyen
M.,
Osipo
C.,
Targeting Breast Cancer Stem Cells Using Naturally Occurring Phytoestrogens. Int J Mol Sci.
2022;
23
(12)
:
6813
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 12 No 1 (2025)
Page No.: 7097-7108
Published on: 2025-01-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 1181 times
- PDF downloaded - 372 times
- XML downloaded - 97 times
 Biomedpress
Biomedpress







