Abstract
Background: Multiple sclerosis (MS) is an autoimmune disease characterized by chronic inflammation in the central nervous system (CNS). MicroRNAs (miRNAs) are tiny molecules that act as regulators within cells, influencing various processes linked to diseases like MS. Understanding the specific role of miRNAs in MS is crucial for developing new treatment strategies. This study focused on an Iranian population with relapsing-remitting MS. The researchers aimed to examine the levels of a particular miRNA, miR-485-3p, and its target gene, HLADRB1, over a minimum two-month period. By investigating these molecules, the study sought to shed light on the potential involvement of miR-485-3p in the pathogenesis of MS.
Methods: This study investigated the relationship between miR-485-3p and relapsing-remitting multiple sclerosis (RRMS) using a case-control design. The researchers analyzed the expression levels of both miR-485-3p and its potential target gene, HLADRB1, in peripheral blood mononuclear cells (PBMCs) collected from participants. The study included 90 individuals: 30 diagnosed with RRMS who were experiencing a relapse, 30 diagnosed with RRMS who had been in a relapse state for at least two months, and a control group of 30 healthy subjects.
Results: The expression of miR-485-3p was different in the two groups studied (P < 0.0002 and P < 0.001, respectively). RRMS patients in relapse and those at least two months post-relapse showed increased expression compared to the normal group. Additionally, we found increased expression of HLADRB1 in RRMS patients compared to healthy control subjects (P < 0.0001 and P < 0.0003, respectively).
Conclusion: According to the study's findings, miR-485-3p, at least in the Iranian population studied, is likely to be an important biomarker for the early diagnosis of RRMS. However, HLADRB1 might be a crucial target for the development of this illness. Nevertheless, more research is required to provide a definitive answer.
Introduction
Multiple sclerosis (MS) is an autoimmune disease characterized by chronic inflammation in the central nervous system (CNS), the control center of our body. The destruction of axons, damage to myelin, progressive exacerbation, relapses, and recovery are some of the main characteristics of this disease1, 2, 3. It is estimated that up to two million people worldwide are affected by this disease. The disease usually occurs in people in their thirties and forties, and women are diagnosed about three times as often as men4, 5, 6. Studies show that MS occurrence is increasing in developing countries; in Isfahan, one of the largest cities in Iran, the prevalence is 85.8 per 100,000 people7. MS can affect people of all ages and genders, but it affects more women aged 20 - 40. This autoimmune disease is usually relapsing, with relapses alternating with remissions. Relapsing-remitting MS (RRMS) is the most common form, affecting around 85% of MS patients. It's characterized by phases of worsening symptoms (relapses) followed by recovery phases (remissions). Secondary progressive MS (SPMS) develops from RRMS, in which the relapses become less frequent, but the symptoms continue to worsen over time. Primary progressive MS (PPMS) is rarer than RRMS, with gradual worsening of symptoms from the beginning, without pronounced relapses or remissions. Progressive-relapsing MS (PRMS) is the least common form, characterized by a steady progression of symptoms with occasional flare-ups of new or worsening symptoms8, 9, 10.
The most prevalent type of MS is RRMS, characterized by flare-ups and remissions in symptoms11. The course of the disease is often unpredictable, and the severity of symptoms can vary greatly. These symptoms can affect the ability to move, feel, and think. The lack of a standardized detection method complicates the diagnosis of active MS. Recently, promising progress has been made in identifying the genetic factors associated with the development of MS12, 13, 14, 15. A class of short, non-coding RNAs with an average length of 22 nucleotides is referred to as miRNAs. miRNAs are essential for various biological functions, such as differentiation, proliferation, and programmed cell death16, 17, 18, 19, 20.
MS may be associated with abnormal miRNAs that malfunction in immune cells of the blood and glial cells of the CNS. These abnormal miRNAs are likely responsible for the immune system abnormalities observed in MS patients. Studies have gradually shown that changes in the patterns of miRNA expression in the immune cells and brain tissue of MS patients are linked to the progression of the disease21, 22, 23. The exact role and underlying mechanisms of miRNAs in MS are still unclear. In this study, a database called miRWalk was used to identify miR-485-3p as a potentially important miRNA in MS. This suggests that miR-485-3p may play a crucial role in the development of MS. Research into the role of miRNAs in MS paves the way for the development of new miRNA-based therapies. This study, therefore, investigated whether changes in the levels of miR-485-3p and HLA-DRB1 could serve as diagnostic markers for patients with RRMS.
Methods
Study Population
In this case-control study, blood samples from two groups were compared. A total of 60 RRMS patients were included; half (30) suffered relapses, while the other half (30) had persistent relapses for at least two months. Healthy controls: 30 healthy individuals were recruited from the MS research center in Isfahan. These individuals matched the patient group in terms of age and gender and had no history of autoimmune diseases, which was confirmed by a medical examination. All MS diagnoses were confirmed by a neurology expert using the established McDonald diagnostic criteria17. To ensure medications wouldn't confound the results, the researchers only included patients with recurrent disease who hadn't undergone any treatment for at least two months before the study began. Within this group, they specifically selected patients who had been treated only with beta-interferon (IFN-β) for at least two months prior to disease recurrence. All subjects provided their informed consent, and the study was ethically approved by the Islamic Azad University's Ashkezar Branch Ethics Committee. For the analysis of miR-485-3p expression, the researchers collected 4 ml of peripheral blood in EDTA tubes. These blood samples were stored on ice during transportation to the laboratory for analysis by qRT-PCR.
PBMCs Isolation
This study isolated peripheral blood mononuclear cells (PBMCs) from blood samples using a density gradient centrifugation technique, following the manufacturer's instructions for Lymphoprep. Here's a breakdown of the process: (a) Blood Dilution and Layering: Four milliliters of blood were first diluted 1:1 with physiological saline. This diluted blood was then carefully layered on top of a 4 ml volume of Lymphoprep solution in a Falcon tube. (b) Centrifugation and PBMC Collection: The layered Falcon tubes were centrifuged at 800 g for 30 minutes. This centrifugation process separates the blood components based on their density. PBMCs, located in the middle layer, were then carefully transferred to a separate, RNase-free microtube with a capacity of 2 ml. (c) Storage: Finally, the collected PBMCs were frozen at -20°C for later analysis.
RNA Isolation, cDNA Synthesis, and RT-qPCR
Firstly, RNA was isolated from samples using the RNA hybrid R kit. The quality and quantity of the extracted RNA were assessed using gel electrophoresis and NanoDrop. To synthesize cDNA for further analysis, two approaches were used: miR-485-3p: A standard cDNA synthesis kit was employed following the manufacturer's instructions. HLADRB1: cDNA synthesis was performed directly on the total RNA using a separate cDNA synthesis kit according to its specific protocol. The Rotor-Gene 6000 system was used for RT-qPCR; the assay was performed following the instructions. U6 and GAPDH were used as housekeeping genes for analyzing the expression change of HLADRB1 and miR-485-3p. Data analysis was performed using the 2-ΔΔCT method.
Statistical Analysis
The GraphPad Prism software (GraphPad, USA, version 5.01) was used for the statistical analysis. The Kolmogorov–Smirnov test, a nonparametric test, was performed to determine normality, and ANOVA was used to analyze data between groups. For all tests, p ≤ 0.05 was set as the significance level.
| Characteristics | Control (N = 30) | Recurring patients (N = 30) | Two months after relapse patients (N = 30) |
| Sex | |||
| Number of males | 13 | 11 | 10 |
| Number of females | 17 | 19 | 20 |
| Mean age | 33.72 ± 6.12 | 32.28 ± 2.58 | 34.54 ± 1.25 |
| Mean of disease duration (years) | - | 7.63 ± 0.72 | 6.25 ± 1.82 |
| Family history | - | 9 | 12 |
| Drug: Interferon | - | 22 | 16 |
Results
Characteristics of Study Population
A total of 90 participants were enrolled in the present study, including 60 RRMS patients. Among them, 30 had a recurrence (mean age: 32.28 ± 2.58 years, 11 men and 19 women) and 30 patients who had experienced a recurrence for at least two months (mean age: 34.54 ± 1.25 years, 10 men and 20 women), age- and sex-matched healthy individuals, and 30 healthy subjects (mean age: 33.72 ± 6.12 years, 13 men and 17 women). There was no significant correlation between men and women (p-value = 0.28). The biological characteristics of the patients (with recurrences and recurrences lasting for at least two months) and the healthy subjects are listed in Table 1.
Analysis of miR-485-3p Expression
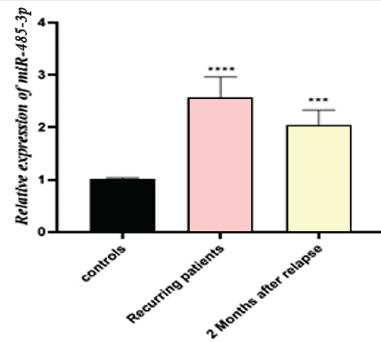
Our research has shown that the relative quantification (RQ) was different across the groups. In our data analysis, there was a significant increase in the expression of miR-485-3p in RRMS patients in comparison to control subjects (P < 0.0002 and P < 0.001, respectively) (Figure 1). Thus, our results suggest that miR-485-3p could be valuable as a novel biomarker for RRMS patients.
Analysis of the Expression Level of miR-485-3p’s Target
The HLADRB1 gene, which is involved in MS and for miR-485-3p according to the miRWalk 2.0 database, was investigated in patients with relapsing and recurrent disease for at least two months compared to control subjects. Our results showed an increased expression of HLADRB1 in patients with recurrent and at least two-month relapses compared to healthy individuals (P = 0.0001 and P = 0.0003, respectively) (Figure 2).
Discussion
In this study, miR-485-3p was nominated as a miRNA intricately involved in RRMS disease. Subsequently, the expression of miR-485-3p was analyzed by real-time quantitative PCR in two groups: RRMS patients (recurrent and relapsed for at least two months) (n = 60) and healthy individuals (n = 30). Data analysis revealed an increased expression of miR-485-3p in patients with recurrent disease and relapses of at least two months compared to controls. Considering that the most ideal method to investigate the role of miR-485-3p in RRMS is to collect samples from nervous system tissue, we used PBMC samples due to the difficulty of sample collection and the utilization of the biomarker potential of miR-485-3p. We also found that the expression of HLA-DRB1 was significantly increased in RRMS patients compared to controls.
We postulated that, in light of our observations, the overexpression of miR-485-3p in the RRMS group relative to the control group may one day be studied as a possible target for therapy. The course of a patient's MS might vary; months after the disease first manifests, they typically continue to experience neurological impairment and clinical disease activity. For patient care, biomarkers that indicate the likelihood of a therapeutic response are ideal24, 25, 26. Several studies have shown the correlations between the progression of MS and the aberrant expression of miRNAs27, 28, 29. The results of the differential expression study in RRMS samples revealed the overexpression of eight out of nine highly dysregulated miRNAs, including miR-485-3p30. TaqMan array analysis showed that miR-485-3p was significantly increased in CD4+ T cells from peripheral blood samples of RRMS patients31. Ten miRNAs, including miR-485-3p, were identified in miRNA profiling studies in CD4+ T cells from RRMS patients using TLDA32. The outcomes of earlier research support our findings and corroborate the conclusions of our investigation.
There is sufficient evidence for the role of HLA II in MS. HLA-DRB1 expression on the cell surface has specifically increased, according to flow cytometric analysis33, 34. One of the most crucial requirements for creating a miRNA-based biomarker for MS, or any other disease, is the capacity to quantify miRNAs from a range of samples with adequate sensitivity, precision, and repeatability. Gaining insight into the intricacy of miRNAs may provide new opportunities for identifying personalized biomarkers for clinical diagnosis and tracking treatment effectiveness. At the very least, for the Iranian population, miR-485-3p may prove to be a useful biomarker and molecular target in the future when devising novel approaches to manage RRMS. Since miR-485-3p likely regulates multiple genes, it might play a diverse range of functions in biological processes related to MS. This opens exciting avenues for future research to explore these additional roles. This study highlights the potential significance of miR-485-3p and HLA-DRB1 in MS. They could potentially serve as biomarkers for early diagnosis of RRMS or as targets for developing new treatment strategies. However, further studies are crucial to definitively establish their exact role in MS.
Conclusions
The results indicate that the deregulation of miR-485-3p is likely associated with RRMS and poor prognosis and may have functional significance through its target. This miRNA and its target could potentially serve as valuable biomarkers for the early diagnosis of RRMS and the prediction of treatment response. Identifying genetic factors involved in MS could contribute to a better understanding of the pathophysiology, prognosis, and treatment of this disease.
Abbreviations
MS - Multiple Sclerosis, CNS - Central Nervous System, miRNA - MicroRNA, RRMS - Relapsing-Remitting Multiple Sclerosis, PBMCs - Peripheral Blood Mononuclear Cells, SPMS - Secondary Progressive Multiple Sclerosis, PPMS - Primary Progressive Multiple Sclerosis, PRMS - Progressive-Relapsing Multiple Sclerosis, IFN-β - Interferon beta, EDTA - Ethylenediaminetetraacetic acid, qRT-PCR - Quantitative Real-Time Polymerase Chain Reaction, cDNA - Complementary DNA, RQ - Relative Quantification, HLA - Human Leukocyte Antigen
Acknowledgments
The authors would like to appreciate and thank all those involved in this project.
Author’s contributions
All authors equally contributed to this work, read and approved the final manuscript.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All subjects provided their informed consent, and the study was approved ethically by the Islamic Azad University's Ashkezar Branch Ethics Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Ransohoff
R.M.,
Hafler
D.A.,
Lucchinetti
C.F.,
Multiple sclerosis-a quiet revolution. Nature Reviews. Neurology.
2015;
11
(3)
:
134-42
.
View Article PubMed Google Scholar -
Lin
X.,
Wang
S.,
Gao
Y.,
The effects of intermittent fasting for patients with multiple sclerosis (MS): a systematic review. Frontiers in Nutrition.
2024;
10
:
1328426
.
View Article PubMed Google Scholar -
Hoseini
S.H.,
Enayati
P.,
Nazari
M.,
Babakhanzadeh
E.,
Rastgoo
M.,
Sohrabi
N.B.,
Biomarker Profile of Colorectal Cancer: Current Findings and Future Perspective. Journal of Gastrointestinal Cancer.
2024;
55
(2)
:
497-510
.
View Article PubMed Google Scholar -
Al-Kuraishy
H.M.,
Jabir
M.S.,
Al-Gareeb
A.I.,
Saad
H.M.,
Batiha
G.E.,
Klionsky
D.J.,
The beneficial role of autophagy in multiple sclerosis: yes or No?. Autophagy.
2024;
20
(2)
:
259-74
.
View Article PubMed Google Scholar -
Stefanou
M.I.,
Giannopapas
V.,
Kitsos
D.K.,
Chondrogianni
M.,
Theodorou
A.,
Kosmidou
M.,
Prevalence and epidemiology of stroke in patients with multiple sclerosis: a systematic review and meta-analysis. Journal of Neurology.
2024;
271
(7)
:
4075-85
.
View Article PubMed Google Scholar -
Jahangir
M.,
Nazari
M.,
Babakhanzadeh
E.,
Manshadi
S.D.,
Where do obesity and male infertility collide?. BMC Medical Genomics.
2024;
17
(1)
:
128
.
View Article PubMed Google Scholar -
Barzegar
M.,
Vaheb
S.,
Mirmosayyeb
O.,
Ashtari
F.,
Afshari-Safavi
A.,
Adibi
I.,
Prevalence and incidence of multiple sclerosis in Isfahan, Iran between 1996 and 2021: A population-based study. Multiple Sclerosis and Related Disorders.
2024;
84
:
105479
.
View Article PubMed Google Scholar -
Goldenberg
M.M.,
Multiple sclerosis review. Pharmacy and therapeutics.
2012;
37
(3)
:
175
.
PubMed Google Scholar -
Gajewski
B.,
Karlińska
I.,
Stasio\lek
M.,
Symbol Digit Modalities Test in progressive multiple sclerosis. Neurologia i Neurochirurgia Polska.
2024;
58
(3)
:
221-32
.
View Article PubMed Google Scholar -
Huang
C.,
Esfani Sarafraz
P.,
Enayati
P.,
Mortazavi Mamaghani
E.,
Babakhanzadeh
E.,
Nazari
M.,
Circular RNAs in renal cell carcinoma: from mechanistic to clinical perspective. Cancer Cell International.
2023;
23
(1)
:
288
.
View Article PubMed Google Scholar -
Iaffaldano
P.,
Lucisano
G.,
Guerra
T.,
Patti
F.,
Cocco
E.,
De Luca
G.,
Italian MS Register
Evaluation of drivers of treatment switch in relapsing multiple sclerosis: a study from the Italian MS Registry. Journal of Neurology.
2024;
271
(3)
:
1150-9
.
View Article PubMed Google Scholar -
Ascherio
A.,
Munger
K.L.,
Environmental risk factors for multiple sclerosis. Part II: noninfectious factors. Annals of Neurology.
2007;
61
(6)
:
504-13
.
View Article PubMed Google Scholar -
Stangel
M.,
Penner
I.K.,
Kallmann
B.A.,
Lukas
C.,
Kieseier
B.C.,
Towards the implementation of `no evidence of disease activity' in multiple sclerosis treatment: the multiple sclerosis decision model. Therapeutic Advances in Neurological Disorders.
2015;
8
(1)
:
3-13
.
View Article PubMed Google Scholar -
Sawcer
S.,
Franklin
R.J.,
Ban
M.,
Multiple sclerosis genetics. Lancet Neurology.
2014;
13
(7)
:
700-9
.
View Article PubMed Google Scholar -
Alipourfard
I.,
Khorshidian
A.,
Babakhanzadeh
E.,
Nazari
M.,
Susceptibility to azoospermia by haplotype analysis of protamine 1 and protamine 2 variants. Human Gene.
2023;
37
:
201200
.
View Article Google Scholar -
Ogata-Kawata
H.,
Izumiya
M.,
Kurioka
D.,
Honma
Y.,
Yamada
Y.,
Furuta
K.,
Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One.
2014;
9
(4)
:
e92921
.
View Article PubMed Google Scholar -
Witwer
K.W.,
Circulating microRNA biomarker studies: pitfalls and potential solutions. Clinical Chemistry.
2015;
61
(1)
:
56-63
.
View Article PubMed Google Scholar -
Barbarotto
E.,
Schmittgen
T.D.,
Calin
G.A.,
MicroRNAs and cancer: profile, profile, profile. International Journal of Cancer.
2008;
122
(5)
:
969-77
.
View Article PubMed Google Scholar -
Almeida
M.I.,
Reis
R.M.,
Calin
G.A.,
MicroRNA history: discovery, recent applications, and next frontiers. Mutation Research.
2011;
717
(1-2)
:
1-8
.
View Article PubMed Google Scholar -
Nazari
M.,
Khorshidian
A.,
Alizadeh
S.,
Falahati
A.M.,
Haghparast
A.,
Ghasemifar
S.,
Association between peroxisome proliferator activated receptor gamma coactivator 1 gene with overweight and obesity risk: case-control study and meta-analysis. Human Gene.
2022;
34
:
201123
.
View Article Google Scholar -
Arruda
L.C.,
Lorenzi
J.C.,
Sousa
A.P.,
Zanette
D.L.,
Palma
P.V.,
Panepucci
R.A.,
Autologous hematopoietic SCT normalizes miR-16, -155 and -142-3p expression in multiple sclerosis patients. Bone Marrow Transplantation.
2015;
50
(3)
:
380-9
.
View Article PubMed Google Scholar -
Miyazaki
Y.,
Li
R.,
Rezk
A.,
Misirliyan
H.,
Moore
C.,
Farooqi
N.,
Autoimmunity
CIHR/MSSC New Emerging Team Grant in Clinical,
Team
MSSRF Canadian B cells in MS,
A novel microRNA-132-sirtuin-1 axis underlies aberrant B-cell cytokine regulation in patients with relapsing-remitting multiple sclerosis [corrected]. PLoS One.
2014;
9
(8)
:
e105421
.
View Article PubMed Google Scholar -
Babakhanzadeh
E.,
Danaei
H.,
Abedinzadeh
M.,
Ashrafzadeh
H.R.,
Ghasemi
N.,
Association of miR-146a and miR196a2 genotype with susceptibility to idiopathic recurrent pregnancy loss in Iranian women: A case-control study. International Journal of Reproductive Biomedicine.
2021;
19
(8)
:
725-32
.
View Article PubMed Google Scholar -
Disanto
G.,
Barro
C.,
Benkert
P.,
Naegelin
Y.,
Schädelin
S.,
Giardiello
A.,
Swiss Multiple Sclerosis Cohort Study Group
Serum Neurofilament light: A biomarker of neuronal damage in multiple sclerosis. Annals of Neurology.
2017;
81
(6)
:
857-70
.
View Article PubMed Google Scholar -
Hauser
S.L.,
Bar-Or
A.,
Comi
G.,
Giovannoni
G.,
Hartung
H.P.,
Hemmer
B.,
I
OPERA,
Clinical Investigators
OPERA II,
Ocrelizumab versus interferon beta-1a in relapsing multiple sclerosis. The New England Journal of Medicine.
2017;
376
(3)
:
221-34
.
View Article PubMed Google Scholar -
Khodadadian
A.,
Varghaiyan
Y.,
Babakhanzadeh
E.,
Alipourfard
I.,
Haghi-Daredeh
S.,
Ghobadi
A.,
Fertility preservation in women with ovarian cancer: Finding new pathways: A case-control study. International Journal of Reproductive Biomedicine.
2021;
19
(2)
:
157-66
.
View Article PubMed Google Scholar -
Vistbakka
J.,
Elovaara
I.,
Lehtimäki
T.,
Hagman
S.,
Circulating microRNAs as biomarkers in progressive multiple sclerosis. Multiple Sclerosis.
2017;
23
(3)
:
403-12
.
View Article PubMed Google Scholar -
Regev
K.,
Healy
B.C.,
Khalid
F.,
Paul
A.,
Chu
R.,
Tauhid
S.,
Association between serum microRNAs and magnetic resonance imaging measures of multiple sclerosis severity. JAMA Neurology.
2017;
74
(3)
:
275-85
.
View Article PubMed Google Scholar -
Nazari
M.,
Babakhanzadeh
E.,
Mohsen Aghaei Zarch
S.,
Talebi
M.,
Narimani
N.,
Dargahi
M.,
Upregulation of the RNF8 gene can predict the presence of sperm in azoospermic individuals. Clinical and Experimental Reproductive Medicine.
2020;
47
(1)
:
61-7
.
View Article PubMed Google Scholar -
Güllüoğlu
H.,
Uysal
H.,
Poyraz
T.,
Altun
Z.,
Kaya
D.,
Özçelik
P.,
Differences in the Differential Expression of MicroRNAs Between Patients with Familial Multiple Sclerosis and Those with Sporadic Multiple Sclerosis. Meandros Medical and Dental Journal.
2023;
24
(4)
:
334-42
.
View Article Google Scholar -
Jiang-xiu
C.,
Shi-song
M.,
Zhu
H.,
Diagnostic value of miR-485-3p in IgA nephropathy and its correlation with immunological parameters and T lymphocyte subsets. Journal of Clinical Nephrology..
2022;
22
(11)
:
924-30
.
-
Xue
Y.,
Zhang
L.,
Guo
R.,
Shao
X.,
Shi
M.,
Yuan
C.,
miR-485 regulates Th17 generation and pathogenesis in experimental autoimmune encephalomyelitis through targeting STAT3. Journal of Neuroimmunology.
2023;
379
:
578100
.
View Article PubMed Google Scholar -
Silvestri
A. De,
Capittini
C.,
Mallucci
G.,
Bergamaschi
R.,
Rebuffi
C.,
Pasi
A.,
The involvement of HLA class II alleles in multiple sclerosis: a systematic review with meta-analysis. Disease Markers.
2019;
2019
(1)
:
1409069
.
View Article PubMed Google Scholar -
Michalik
J.,
\vCierny
D.,
Kantorová
E.,
Kantárová
D.,
Juraj
J.,
Párnická
Z.,
The association of HLA-DRB1 and HLA-DQB1 alleles with genetic susceptibility to multiple sclerosis in the Slovak population. Neurological Research.
2015;
37
(12)
:
1060-7
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 11 No 10 (2024)
Page No.: 6825-6830
Published on: 2024-10-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
- HTML viewed - 1488 times
- XML downloaded - 63 times
- PDF downloaded - 527 times
 Biomedpress
Biomedpress



