Abstract
Introduction: This study aimed to compare the efficacy of Copy Number Variation sequencing (CNV-Seq) with that of traditional karyotyping in prenatal diagnostics by assessing their concordance and ability to identify aneuploidies and structural abnormalities in fetal chromosomes.
Methodology: We analyzed 177 amniotic fluid samples from pregnant women who were at or beyond 16 weeks of gestation, utilizing both CNV-Seq and karyotyping to evaluate their detection capabilities.
Results: CNV-Seq identified chromosomal abnormalities in 46 cases (26.0%), demonstrating a higher detection rate compared to karyotyping, which found abnormalities in 40 cases (22.6%). CNV-Seq showed 100% concordance in identifying conditions such as trisomy 21, 18, 13, monosomy X, and 47, XXY. It also detected three mosaic cases and 13 copy number variations (CNVs) involving deletions or duplications that were not fully concordant with karyotyping results. Notably, CNV-Seq had a detection rate of 3.95% (7/177) for pathogenic or likely pathogenic chromosomal anomalies, and variants of uncertain significance (VUS) constituted 3.39% (6/177) of the findings.
Conclusion: CNV-Seq improves the precision of prenatal diagnostics and broadens the scope for informed clinical decision-making, especially in managing pregnancies with detected abnormalities. The integration of CNV-Seq with traditional karyotyping addresses gaps in detection and supports a more comprehensive approach to prenatal care. Further studies should aim to include a broader and more diverse population to validate and expand upon these results.
Introduction
The emergence of genomic technologies has dramatically revolutionized the field of prenatal diagnostics, significantly advancing our comprehension of fetal chromosomal abnormalities. Whereas traditional karyotyping has played a pivotal role in identifying aneuploidies and significant chromosomal rearrangements, it is hindered by its low resolution and the protracted periods required for cell culture1, 2, 3, 4. These constraints are particularly critical in contexts where prompt and accurate prenatal decision-making is paramount. To address these limitations, sophisticated methodologies such as Copy Number Variation sequencing (CNV-Seq) have been introduced, providing expedited and exhaustive genomic analyses4, 5.
CNV-Seq is superior in detecting submicroscopic chromosomal anomalies that remain undetected by traditional karyotyping, thereby significantly augmenting the diagnostic yield for chromosomal abnormalities6, 7, 8, 9. Luo et al. (2023) conducted a meta-analysis of eight studies, encompassing 11,091 pregnant women identified as high-risk or bearing fetuses with structural abnormalities detected through ultrasound. This systematic review demonstrated that CNV-Seq uncovered an additional 2% (95% CI, 0% to 4%) of chromosomal anomalies beyond what was detected by traditional karyotyping across six series. Moreover, a pooled mean incremental yield of 4% (95% CI, 3%–6%) in pathogenic CNVs was reported, with a range spanning 1%–16%. The findings underscored the enhanced capability of CNV-Seq in prenatal diagnosis, highlighting its expansive coverage, high throughput, elevated resolution, culture-independent process, excellent compatibility, and adjustable sequencing depth as factors contributing to its significant value in prenatal diagnostics10.
Nevertheless, CNV-Seq presents its own set of challenges. Although it can identify copy number variations at the kilobase level, it is incapable of detecting balanced translocations and inversions that do not result in copy number changes. Therefore, interpreting results from CNV-Seq necessitates a sophisticated understanding of genomic contexts and often requires the complementary use of traditional diagnostic methods to achieve a thorough genetic evaluation10, 11, 12, 13.
The Clinical Genome Resource (ClinGen) and the American College of Medical Genetics and Genomics (ACMG) have issued guidelines to classify CNVs based on their pathogenicity, ranging from benign to pathogenic4. These classifications assist clinicians in making well-informed decisions; however, the variability in the expressivity and penetrance of CNVs presents considerable challenges in counseling14, 15, 16. The detection of variants of uncertain significance further emphasizes the importance of a meticulous correlation between genotype and phenotype, a task complicated by the wide spectrum of clinical manifestations associated with CNVs17, 18, 19.
In Vietnam, where congenital anomalies considerably affect neonatal health, adopting CNV-Seq in prenatal care could potentially diminish the incidence of genetic disorders and enhance pregnancy outcomes. Annually, approximately 40,000 newborns in Vietnam suffer from congenital anomalies, constituting 1.5-2% of births. Given the lack of specific treatments for most of these anomalies, prenatal screening and diagnosis become crucial for genetic counseling, pregnancy management, and postnatal care20, 21, 22.
Our investigation aims to assess the efficacy of CNV-Seq relative to traditional karyotyping within a clinical context, evaluating their concordance and the incremental diagnostic value CNV-Seq might offer. By analyzing amniotic fluid samples from pregnant women at or beyond a gestational age of 16 weeks, we seek to elucidate the implications of CNV-Seq for prenatal diagnosis, focusing on its impact on clinical practice and genetic counseling.
Methods
Study Design and Participant Overview
Study Setting and Duration
The study was conducted at a specialized Center for Prenatal Screening, Diagnosis, and Neonatology in a major regional hospital, equipped with state-of-the-art genetic screening technology. It commenced in January 2021 and concluded in December 2022, aiming to collect a comprehensive dataset throughout different seasons.
Participants
Eligible participants were pregnant women who were at least 16 weeks into their gestation, deemed an optimal period for amniocentesis, which ensures the availability of sufficient amniotic fluid for genetic testing. Enrollment was limited to those with singleton pregnancies to reduce genetic variability and exclude the confounding variables associated with multiples. Inclusion criteria included elevated risk for chromosomal abnormalities indicated by abnormal prenatal screening results, concerning ultrasound findings, or a family history of genetic disorders. Exclusion criteria encompassed incomplete data for CNV-Seq and karyotyping and multiple pregnancies, maintaining the genetic analysis's focus and integrity.
Procedures and Genetic Analysis
Amniocentesis and Sample Preparation
Amniocentesis was carried out between 16 and 18 weeks of gestation under ultrasound guidance, withdrawing approximately 20-30 mL of amniotic fluid. This timeframe minimizes risk to both the mother and fetus while providing an adequate sample volume for genetic testing.
Karyotyping
Approximately 10 mL of the collected fluid was treated with colchicine, subjected to hypotonic treatment, fixation, and centrifugation. Chromosomes were stained using G-banding techniques and analyzed with the GSL-120 automatic metaphase chromosome analysis system (Leica Microsystems, Deerfield, IL, USA), with chromosomal structures cataloged according to the 2016 International System of Human Cytogenetic Nomenclature23, 24.
CNV-Sequencing (CNV-seq)
The leftover sample was allocated for CNV-sequencing by Berry Genomics Co., including quality control measures like the usage of STR markers to ensure genetic integrity and prevent maternal DNA contamination. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA), prepared into a sequencing library with the NEBNext Ultra II DNA Library Prep Kit for Illumina, and sequenced on the NextSeq 500 platform (Illumina, San Diego, CA, USA). The produced data were aligned to human genome references hg19 and updated to hg38 (GRCh38) using the DECIPHER database, with CNVs interpreted in alignment with public databases and evaluated according to ACMG guidelines for clinical relevance.
Data Collection and Statistical Analysis
Data Collection and Management
Clinical and genetic data collection adhered to standardized protocols for accuracy and reliability. High-resolution ultrasound results and genetic test findings from karyotyping and CNV-sequencing were meticulously recorded and entered into an electronic database. Genetic specialists reviewed the data, cross-referencing each finding with reputable genomic databases like DECIPHER and OMIM to guarantee consistent and reliable data interpretation.
Statistical Analysis
The statistical analysis aimed to accurately represent the data, focusing on demographic characteristics and chromosomal abnormality incidences. Data management and analysis were conducted using IBM SPSS Statistics (version 22.0, IBM Corp., Armonk, NY, USA), STATA (version 17.0, StataCorp LLC, College Station, TX, USA), and R software (version 4.3.2), calculating means, standard deviations, and percentages for maternal and gestational ages at diagnosis. Genetic findings were categorized and quantitatively expressed as percentages, including normal findings, duplications, deletions, mosaicism, and aneuploidy.
Ethical Considerations
The study was approved by the Institutional Review Board of Nghe An Maternity and Pediatric Hospital (Code 13/QĐ-BVSN dated 16/01/2023). Informed consent was obtained from all participants, detailing the study's purpose, procedures, and potential risks, ensuring informed decision-making. Participant privacy was safeguarded with unique identification codes, maintaining confidentiality. Amniocentesis was performed by certified professionals following clinical standards to minimize risks, with continuous monitoring to address any complications promptly, ensuring participant safety and study integrity.
| Characteristics | Number (n) | Percentage (%) |
|---|---|---|
| Maternal Age | ||
| < 35 years | 117 | 66.1 |
| ≥ 35 years | 60 | 33.9 |
| Average (years) | Mean ± SD: 31.3 ± 6.9 | Range: 15-47 years |
| Gestational Age | ||
| < 20 weeks | 97 | 54.8 |
| ≥ 20 weeks | 80 | 45.2 |
| Average (weeks) | Mean ± SD: 20.5 ± 4.0 | Range: 16-33 months |
| Indication | Number (n) | Percentage (%) |
|---|---|---|
| Abnormal ultrasound morphology | 120 | 67.8 |
| High-risk maternal serum screening | 19 | 10.7 |
| High-risk NIPT (Non-invasive prenatal test) | 5 | 2.8 |
| History of pregnancies with abnormalities | 4 | 2.3 |
| Abnormal ultrasound + High-risk maternal serum screening | 11 | 6.2 |
| Abnormal ultrasound + High-risk NIPT | 14 | 7.9 |
| Abnormal ultrasound + History of pregnancies with abnormalities | 4 | 2.3 |
| Total | 177 | 100 |
| Results | CNV-seq Number (n) | CNV-seq Percentage (%) | Karyotype Number (n) | Karyotype Percentage (%) |
|---|---|---|---|---|
| Total of abnormal patients | 46 | 26.0% | 40 | 22.6% |
| Aneuploid chromosomal abnormalities | 34 | 34 | ||
| 47,XXXY,+21 | 17 | 17 | ||
| 47,XXXY,+18 | 12 | 12 | ||
| 47,XXXY,+13 | 3 | 3 | ||
| 47,XXX | 1 | 1 | ||
| 45,X | 1 | 1 | ||
| Mosaic cases | 3 | 2 | ||
| Triploidy 69,XXX | 0 | 1 | ||
| Deletions/duplications ≥ 5 Mb | 6 # | 3 | ||
| Deletions/duplications < 5 Mb | 7 # | 0 | ||
| Total of normal patients | 131 | 74.0% | 137 | 77.4% |
| Total | 177 | 100% | 177 | 100% |
| No. | CNV-seq Result | Karyotype Result | Pregnancy Outcome |
|---|---|---|---|
| 1 | dup(8)(p22-p21.3) | 46,XY (normal) | 8 months - Normal development |
| 2 | del(4)(p16.3), dup(17)(q24.3-q25.3) | 46,XY (normal) | Terminated pregnancy due to the risk of Wolf-Hirschhorn syndrome |
| 3 | del(5)(p15.33-p15.31) | 46,XY,del(5)(p15.3) | Terminated pregnancy following the diagnosis of Cri-du-chat syndrome |
| 4 | dup(1)(p31.1) | 46,XX (normal) | Normal development at 2 months |
| 5 | del(6)(q27), dup(19)(q13.33-q13.43) | 46,XY (normal) | Pregnancy terminated at 21 weeks (The fetus presents with hydrocephalus and cerebellar hypoplasia) |
| 6 | del(6)(q27), dup(19)(q13.33-q13.43) | 46,XY (normal) | Intrauterine fetal demise at 37 weeks (Previous pregnancy history of abnormalities detected at 26 weeks) |
| 7 | Suspected mosaic trisomy 12 | 47,XX,i(12)(p10),i(12)(q10)[8%]/46,XX[92%] | Terminated pregnancy due to critical heart defects |
| 8 | No abnormalities detected | 69,XXX | Fetal demise at 20 weeks due to severe growth retardation |
| 9 | del(18)(p11.32), dup(9)(q21.11-q34.3) | 46,XX,+t(9;15)(q21;q2),-15,+t(15;18)(q21;p11.3),-18 | Pregnancy terminated due to severe genetic anomalies |
| 10 | dup(6)(q15-q16.1) | 46,XY (normal) | Normal development at 9 months |
| 11 | dup(18)(p11.32-p11.21): Homozygous | 47,XY,+mar | Normal development at 6 months with 6 fingers on the right hand, cryptorchidism, subclinical hypothyroidism, slight aortic arch stenosis |
| 12 | Suspected mosaic sex chromosome abnormality | 46,XY (normal) | Normal development at 6 months |
| Type of CNV | Pathogenic | Likely pathogenic | Uncertain significance | Total |
|---|---|---|---|---|
| Deletions | 2 | 0 | 3 | 5 |
| Duplications | 1 | 4 | 3 | 8 |
| Total | 3 | 4 | 6 | 13 |
| Detection rate (%) (n = 177) | 1.7% (3/177) | 2.3% (4/177) | 3.9% (6/177) | 7.3% (13/177) |
Results
Characteristics of the Study Group
The study involved a comprehensive analysis of maternal age and gestational age at the time of amniocentesis across a cohort of 177 participants. The distribution of maternal age demonstrated a bimodal pattern, with the majority (66.1%, n=117) of participants under the age of 35. The mean maternal age was 31.3 years (SD ± 6.9; range 15–47 years). The mean gestational age at the time of amniocentesis was 20.5 weeks (SD ± 4.0; range 16–33 weeks), with the majority of procedures (54.8%, n=97) performed before 20 weeks' gestation. These findings are visualized in Figure 1 and Figure 2 and detailed in Table 1.
Indications for Amniotic Fluid Analysis
The primary indication for amniotic fluid testing was abnormal ultrasound morphology, which accounted for 67.8% (120/177) of all cases. Other indications included high-risk maternal serum screening results (10.7%, n=19) and high-risk results from non-invasive prenatal testing (NIPT) (2.8%, n=5). Combined indications included abnormal ultrasound with high-risk maternal serum screening (6.2%, n=11) and abnormal ultrasound with high-risk NIPT results (7.9%, n=14). A history of pregnancies with chromosomal abnormalities was noted in 2.3% (n=4) of cases. These findings are summarized in Table 2.
Chromosomal Abnormalities Detected
In the study cohort of 177 amniotic fluid samples, chromosomal anomalies were identified in 46 cases (26.0%) via CNV-sequencing (CNV-seq), slightly surpassing the detection rate of traditional karyotyping, which identified abnormalities in 40 cases (22.6%). Both diagnostic methods demonstrated a complete concordance rate of 100% in detecting specific aneuploidies: trisomy 21 in 17 cases, trisomy 18 in 12 cases, trisomy 13 in three cases, monosomy X in one case, and 47,XXY in one case.
CNV-seq further identified three cases of mosaicism not initially detected by karyotyping. These included a suspected case of low-level trisomy 18 mosaicism, trisomy 2 mosaicism estimated to affect 10–30% of cells, and a case of sex chromosome mosaicism (60% XY and 40% XX), which likely arose from maternal cell contamination. These findings, confirmed by subsequent karyotyping, highlight the sensitivity of CNV-seq in detecting mosaicism.
Moreover, CNV-seq demonstrated its superior resolution by identifying 13 structural chromosomal changes involving deletions and duplications across nine samples, including several alterations greater than 5 Mb—a refinement not achieved by karyotyping. This capability underscores the effectiveness of CNV-seq in capturing submicroscopic chromosomal changes less than 5 Mb, which traditional methods may overlook. These results are detailed in Table 3, which compares the performance of CNV-sequencing and karyotyping in identifying various chromosomal abnormalities.
Comprehensive Comparative Analysis of CNV-Sequencing and Karyotyping in Prenatal Diagnostics
This section presents a detailed comparative analysis of CNV sequencing (CNV-seq) and traditional karyotyping across twelve cases, showcasing the superior diagnostic capabilities of CNV-seq. Each case provides a factual account of genetic findings and their clinical outcomes, emphasizing the precision of CNV-seq in prenatal diagnostics.
The analysis underscores the diagnostic precision of CNV-seq, particularly in identifying submicroscopic genetic changes, detecting mosaicism, providing precise breakpoints, covering complex abnormalities, and offering comprehensive screening. These attributes demonstrate the comparative advantage of CNV-seq over traditional karyotyping, supporting informed clinical decisions and significantly influencing pregnancy management. Detailed case analyses are presented in Table 4.
Classification of CNV Abnormalities
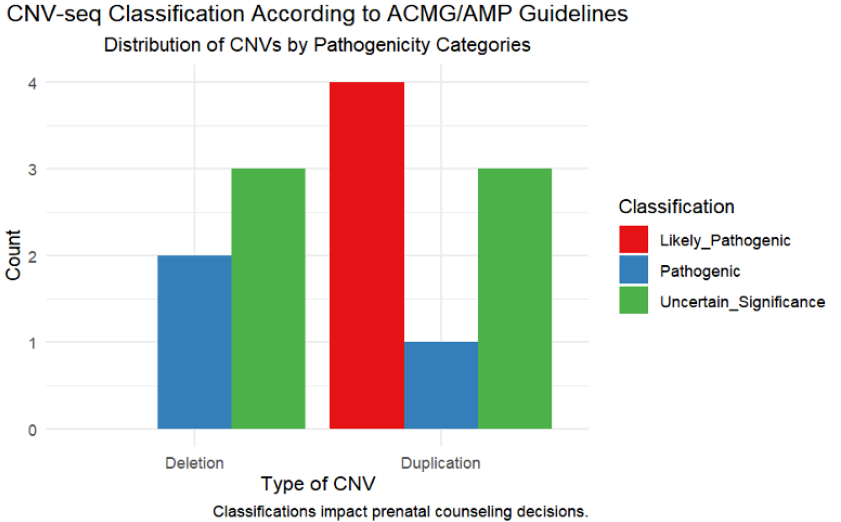
In our study, the classification of CNV abnormalities was performed in strict accordance with the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) guidelines, ensuring the accurate categorization of CNV pathogenicity (refer to Table 5). A total of 13 distinct CNVs, including both deletions and duplications, were identified across nine patients. Out of these, seven were classified as pathogenic or likely pathogenic, underscoring their significant potential for clinical impact. Specifically, our analysis recognized two deletions and one duplication as pathogenic, and four duplications as likely pathogenic. The remaining six CNVs were categorized as variants of uncertain significance (VUS), indicating a more ambiguous potential impact.
Impact on Prenatal Counseling and Clinical Decisions
The pathogenic classification of CNVs, particularly those identified alongside severe morphological anomalies via ultrasound, often guides expectant families toward considering pregnancy termination. This decision is influenced by concerns over the anticipated quality of life and potential severe health challenges for the child. Conversely, CNVs categorized as having uncertain significance pose substantial challenges in prenatal counseling, necessitating a nuanced approach to risk assessment to prevent undue anxiety among prospective parents. Our approach to counseling emphasizes an informed and empathetic methodology, tailored to each case's unique circumstances, ensuring that decisions are made with a comprehensive understanding of all potential outcomes. The impact of CNV classifications on prenatal counseling decisions is depicted in Figure 3.
Discussion
Our study evaluated the diagnostic performance of CNV-Seq compared to traditional karyotyping in detecting chromosomal abnormalities in 177 amniotic fluid samples. CNV-Seq demonstrated a superior detection rate of 26.0% versus 22.6% for karyotyping, including a 3.4% increase in the identification of submicroscopic abnormalities. The technology showed 100% concordance in identifying common aneuploidies and provided additional insights into mosaicism and CNVs not detected by karyotyping. These findings suggest that integrating CNV-Seq into prenatal screening protocols could enhance genetic assessment and improve clinical outcomes, particularly in settings with high incidences of congenital anomalies.
Characteristics of the Participants
Our study included 177 participants, highlighting a trend toward delayed childbearing with a mean maternal age of 31.3 years, reflecting broader implications for prenatal diagnostic strategies. This age profile is slightly older compared to findings from Guangdong, China, where the mean maternal age was 29.84 years11, yet similar to data from Yunnan, China, with ages ranging from 27 to 36 years25. The increased risk of chromosomal anomalies associated with advanced maternal age was significant, demonstrating a higher incidence of chromosome number abnormalities as maternal age increased (p<0.001). However, no significant trends were noted for structural chromosomal abnormalities26, 27, 28. Additionally, the rapid results delivery by CNV-sequencing offers crucial benefits for timely and informed clinical decision-making, especially crucial when anomalies are detected at advanced gestational stages28.
Detection of Genetic Abnormalities by CNV-Sequencing
CNV-Sequencing has proven to be a significant advancement over traditional karyotyping, detecting a wider range of submicroscopic deletions and duplications. Our findings demonstrate a 3.4% increase in the detection rate of chromosomal abnormalities with CNV-Seq, identifying abnormalities in 46 cases (26.0%), compared to 22.6% by karyotyping. This capability reflects the diagnostic limitations of traditional methods, especially in identifying smaller genetic changes.
In a comparative context, a study from Guangzhou, China, using different collection methods including chorionic villus sampling and amniocentesis, found abnormalities in 13.47% of cases11. This contrasts with our exclusive use of amniocentesis, collecting 20-30 mL of amniotic fluid, which may explain the differences in detection rates. Furthermore, our CNV-Seq detected a higher incidence of pathogenic microdeletions/duplications (1.7%) compared to a Yunnan study (0.7%), highlighting the impact of sample source differences on detection rates25.
Our study's higher yield of CNVs and detection rates of pathogenic anomalies (3.95%) also compares favorably with a large Sichuan study, which reported lower rates (2.83%) and a smaller proportion of variants of uncertain significance (1.43%)29. These results suggest that the specific clinical indications for our cohort, primarily abnormal ultrasound morphology, contributed to a more targeted and thus higher yield of detectable abnormalities.
Such findings underline the importance of considering the specific clinical and methodological contexts when interpreting comparative studies. The selection bias inherent in focusing on high-risk populations, as in our study, emphasizes the need for careful generalization of findings to broader populations.
Comparison of CNV-Seq and Karyotype Results Detection Rates and Concordance
Our study, alongside those conducted in Jiangsu and Sichuan, uniformly applied amniocentesis, extracting similar quantities of amniotic fluid during the same gestational stages, adhering to protocols typical in China. Notably, the participant profiles in our and the Jiangsu studies were closely matched, involving high-risk pregnancies, unlike the Sichuan study which included low-risk voluntary participants, potentially diluting its detection rates29, 30. This methodological variance underscores the necessity of considering cohort characteristics when evaluating detection capabilities.
Mosaicism Detection and Structural Changes
CNV-Seq identified three cases of mosaicism and 13 structural chromosomal changes in our study, showcasing its superior resolution over traditional karyotyping. These findings align with the Jiangsu and Sichuan studies, where CNV-Seq also revealed higher incidences of mosaicism and complex structural variations29, 30.
Whole Chromosome Aneuploidies
Both our study and the Jiangsu study predominantly detected trisomy 21, with our study identifying it in 17 cases. Similarly, the Sichuan study reported that 52.2% of whole chromosome aneuploidies were trisomy 21, highlighting the consistent detection capabilities of CNV-Seq across diverse study settings29, 30.
Clinical Outcomes and Confirmations
The clinical implications of CNV-Seq findings significantly influenced pregnancy management decisions, particularly regarding the continuation or termination of pregnancies. The Jiangsu study corroborates the utility of integrating CNV-Seq with karyotyping to enhance diagnostic precision, a practice further validated by the Sichuan study’s 100% concordance rate between CNV-Seq diagnoses and confirmatory tests such as QF-PCR, CMA, and FISH. Additionally, the Sichuan study's follow-up parental DNA testing was crucial for comprehensive genetic counseling and managing prenatal outcomes29, 30.
Strengths and Limitations
Our study marks a significant advancement in prenatal diagnostics in Vietnam by integrating CNV-Seq, which has substantially improved the detection of chromosomal anomalies. CNV-Seq excels particularly in identifying submicroscopic deletions and duplications, areas where traditional karyotyping falls short. This capability not only enriches prenatal diagnostic insights but also enhances clinical decision-making, potentially decreasing the prevalence of genetic disorders.
Despite these advancements, the study faces limitations. Primarily, CNV-Seq cannot detect certain structural and numerical chromosomal aberrations such as balanced translocations and polyploidies, underscoring the continued necessity for traditional karyotyping. Additionally, focusing predominantly on a high-risk Vietnamese population may restrict the broader applicability of our findings, potentially introducing a selection bias that inflates the observed detection rates.
A significant aspect of our study involves the interpretation of variants of uncertain significance (VUS), which presents ongoing challenges in prenatal counseling and patient management. The complexity of interpreting VUS necessitates advanced methodological approaches and a dynamic adaptation to evolving genomic sciences. Current interpretative frameworks, although based on the latest guidelines, may not fully capture the rapidly changing landscape of genomic data.
There is a critical need for future studies to include more diverse genetic cohorts, extending research to varied ethnic backgrounds to ensure the generalizability of our findings. Enhancing counseling protocols and developing robust support systems are essential for maximizing the benefits of advanced genomic technologies like CNV-Seq in prenatal diagnostics.
Conclusion
This study establishes CNV-Seq as a more effective tool than traditional karyotyping for detecting submicroscopic chromosomal anomalies, significantly enhancing prenatal diagnostic precision and supporting more informed clinical decisions. Integrating CNV-Seq with karyotyping is crucial, as it fills the detection gaps for structural abnormalities such as balanced translocations and polyploidies.
The identification of variants of uncertain significance (VUS) through CNV-Seq underscores a critical area for future development, necessitating advancements in genetic counseling and interpretative methodologies. Going forward, expanding research to include broader and more diverse populations will be vital to validate and generalize the diagnostic utility of CNV-Seq and improve the management of VUS in diverse clinical settings.
Abbreviations
ACMG - American College of Medical Genetics and Genomics, CMA - Chromosomal Microarray Analysis, CNV - Copy Number Variation, CNV-Seq - Copy Number Variation Sequencing, ClinGen - Clinical Genome Resource, DECIPHER - Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources, FISH - Fluorescence In Situ Hybridization, GSL-120 - (Specific equipment, not an acronym that requires expansion, mentioned in the context of automatic metaphase chromosome analysis system), NIPT - Non-Invasive Prenatal Testing, OMIM - Online Mendelian Inheritance in Man, PCR - Polymerase Chain Reaction, QF-PCR - Quantitative Fluorescence Polymerase Chain Reaction, STR - Short Tandem Repeat, VUS - Variants of Uncertain Significance
Acknowledgments
The authors would like to thank all the patients who participated in the study.
Author’s contributions
HT conceptualized and designed the study. HT, CN, and HN collected and summarized data. HT, CN, HN, and HL were involved in the data interpretation. HT, HL made the first draft of the paper. All authors contributed significantly to the revision of the manuscript. All authors confirm they read and approved the final version.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Shaffer
L.G.,
Bejjani
B.A.,
Medical applications of array CGH and the transformation of clinical cytogenetics. Cytogenetic and Genome Research.
2006;
115
(3-4)
:
303-9
.
View Article PubMed Google Scholar -
Kearney
H.M.,
Thorland
E.C.,
Brown
K.K.,
Quintero-Rivera
F.,
South
S.T.,
Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee
American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genetics in Medicine.
2011;
13
(7)
:
680-5
.
View Article PubMed Google Scholar -
Wojcik
M.H.,
Reimers
R.,
Poorvu
T.,
Agrawal
P.B.,
Genetic diagnosis in the fetus. Journal of Perinatology.
2020;
40
(7)
:
997-1006
.
View Article PubMed Google Scholar -
Riggs
E.R.,
Andersen
E.F.,
Cherry
A.M.,
Kantarci
S.,
Kearney
H.,
Patel
A.,
Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genetics in Medicine.
2020;
22
(2)
:
245-57
.
View Article PubMed Google Scholar -
Bodurtha
J.,
Strauss
J.F.,
Genomics and perinatal care. The New England Journal of Medicine.
2012;
366
(1)
:
64-73
.
View Article PubMed Google Scholar -
Song
T.,
Xu
Y.,
Li
Y.,
Jia
L.,
Zheng
J.,
Dang
Y.,
Detection of submicroscopic chromosomal aberrations by chromosomal microarray analysis for the prenatal diagnosis of central nervous system abnormalities. Journal of Clinical Laboratory Analysis.
2020;
34
(10)
.
View Article PubMed Google Scholar -
Ma
N.,
Xi
H.,
Chen
J.,
Peng
Y.,
Jia
Z.,
Yang
S.,
Integrated CNV-seq, karyotyping and SNP-array analyses for effective prenatal diagnosis of chromosomal mosaicism. BMC Medical Genomics.
2021;
14
(1)
:
56
.
View Article PubMed Google Scholar -
Shao
Y.,
Yang
S.,
Cheng
L.,
Duan
J.,
Li
J.,
Kang
J.,
Identification of chromosomal abnormalities in miscarriages by CNV-Seq. Molecular Cytogenetics.
2024;
17
(1)
:
4
.
View Article PubMed Google Scholar -
Cicatiello
R.,
Pignataro
P.,
Izzo
A.,
Mollo
N.,
Pezone
L.,
Maruotti
G.M.,
Chromosomal Microarray Analysis versus Karyotyping in Fetuses with Increased Nuchal Translucency. Medical Sciences : Open Access Journal.
2019;
7
(3)
:
40
.
View Article PubMed Google Scholar -
Luo
H.,
Wang
Q.,
Fu
D.,
Gao
J.,
Lu
D.,
Additional diagnostic value of CNV-seq over conventional karyotyping in prenatal diagnosis: A systematic review and meta-analysis. Journal of Obstetrics and Gynaecology Research.
2023;
49
(7)
:
1641-50
.
View Article PubMed Google Scholar -
Lan
L.,
She
L.,
Zhang
B.,
He
Y.,
Zheng
Z.,
Prenatal diagnosis of 913 fetuses samples using copy number variation sequencing. The Journal of Gene Medicine.
2021;
23
(5)
.
View Article PubMed Google Scholar -
Zhang
H.,
Xu
Z.,
Chen
Q.,
Chen
H.,
Ding
X.,
Liu
L.,
Comparison of the combined use of CNV-seq and karyotyping or QF-PCR in prenatal diagnosis: a retrospective study. Scientific Reports.
2023;
13
(1)
:
1862
.
View Article PubMed Google Scholar -
Liu
X.,
Liu
S.,
Wang
H.,
Hu
T.,
Potentials and challenges of chromosomal microarray analysis in prenatal diagnosis. Frontiers in Genetics.
2022;
13
.
View Article PubMed Google Scholar -
Nowakowska
B.,
Clinical interpretation of copy number variants in the human genome. Journal of Applied Genetics.
2017;
58
(4)
:
449-57
.
View Article PubMed Google Scholar -
Westerfield
L.,
Darilek
S.,
van den Veyver
I.B.,
Counseling Challenges with Variants of Uncertain Significance and Incidental Findings in Prenatal Genetic Screening and Diagnosis. Journal of Clinical Medicine.
2014;
3
(3)
:
1018-32
.
View Article PubMed Google Scholar -
Rosenfeld
J.A.,
Coe
B.P.,
Eichler
E.E.,
Cuckle
H.,
Shaffer
L.G.,
Estimates of penetrance for recurrent pathogenic copy-number variations. Genetics in Medicine.
2013;
15
(6)
:
478-81
.
View Article PubMed Google Scholar -
Pollen
A.A.,
Kilik
U.,
Lowe
C.B.,
Camp
J.G.,
Human-specific genetics: new tools to explore the molecular and cellular basis of human evolution. Nature Reviews. Genetics.
2023;
24
(10)
:
687-711
.
View Article PubMed Google Scholar -
Pös
O.,
Radvanszky
J.,
Styk
J.,
Pös
Z.,
Buglyó
G.,
Kajsik
M.,
Copy Number Variation: Methods and Clinical Applications. Applied Sciences (Basel, Switzerland).
2021;
11
(2)
:
819
.
View Article Google Scholar -
Drakulic
D.,
Djurovic
S.,
Syed
Y.A.,
Trattaro
S.,
Caporale
N.,
Falk
A.,
Copy number variants (CNVs): a powerful tool for iPSC-based modelling of ASD. Molecular Autism.
2020;
11
(1)
:
42
.
View Article PubMed Google Scholar -
Giang
H.T.,
Bechtold-Dalla Pozza
S.,
Ulrich
S.,
Linh
L.K.,
Tran
H.T.,
Prevalence and Pattern of Congenital Anomalies in a Tertiary Hospital in Central Vietnam. Journal of Tropical Pediatrics.
2020;
66
(2)
:
187-93
.
View Article PubMed Google Scholar -
Hoang
T.,
Nguyen
D.T.,
Nguyen
P.V.,
Tran
D.A.,
Gillerot
Y.,
Reding
R.,
External birth defects in Southern Vietnam: a population-based study at the grassroots level of health care in Binh Thuan Province. BMC Pediatrics.
2013;
13
(1)
:
67
.
View Article PubMed Google Scholar -
VietnamPlus. Prenatal, new-born screening improves population health 2023 [updated March 25, 2023. Available from: https://en.vietnamplus.vn/prenatal-new-born-screening-improves-population-health-post250449.vnp..
.
-
McGowan-Jordan
J.,
Hastings
R.,
Moore
S.,
Re: International System for Human Cytogenetic or Cytogenomic Nomenclature (ISCN): Some Thoughts, by T. Liehr. Cytogenetic and Genome Research.
2021;
161
(5)
:
225-6
.
View Article PubMed Google Scholar -
Liehr
T.,
International System for Human Cytogenetic or Cytogenomic Nomenclature (ISCN): some Thoughts. Cytogenetic and Genome Research.
2021;
161
(5)
:
223-4
.
View Article PubMed Google Scholar -
Zhang
J.,
Tang
X.,
Hu
J.,
He
G.,
Wang
J.,
Zhu
Y.,
Investigation on combined copy number variation sequencing and cytogenetic karyotyping for prenatal diagnosis. BMC Pregnancy and Childbirth.
2021;
21
(1)
:
496
.
View Article PubMed Google Scholar -
Kim
Y.J.,
Lee
J.E.,
Kim
S.H.,
Shim
S.S.,
Cha
D.H.,
Maternal age-specific rates of fetal chromosomal abnormalities in Korean pregnant women of advanced maternal age. Obstetrics {&}amp; Gynecology Science.
2013;
56
(3)
:
160-6
.
View Article PubMed Google Scholar -
Zhang
X.H.,
Qiu
L.Q.,
Ye
Y.H.,
Xu
J.,
Chromosomal abnormalities: subgroup analysis by maternal age and perinatal features in zhejiang province of China, 2011-2015. Italian Journal of Pediatrics.
2017;
43
(1)
:
47
.
View Article PubMed Google Scholar -
Shi
Y.,
Ma
J.,
Xue
Y.,
Wang
J.,
Yu
B.,
Wang
T.,
The assessment of combined karyotype analysis and chromosomal microarray in pregnant women of advanced maternal age: a multicenter study. Annals of Translational Medicine.
2019;
7
(14)
:
318
.
View Article PubMed Google Scholar -
Wang
J.,
Chen
L.,
Zhou
C.,
Wang
L.,
Xie
H.,
Xiao
Y.,
Prospective chromosome analysis of 3429 amniocentesis samples in China using copy number variation sequencing. American journal of obstetrics and gynecology.
2018;
219
(3)
:
287.e1-287.e18
.
View Article Google Scholar -
Zhang
S.,
Xu
Y.,
Lu
D.,
Fu
D.,
Zhao
Y.,
Combined use of karyotyping and copy number variation sequencing technology in prenatal diagnosis. PeerJ.
2022;
10
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 11 No 7 (2024)
Page No.: 6622-6632
Published on: 2024-07-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 2332 times
- PDF downloaded - 822 times
- XML downloaded - 117 times
 Biomedpress
Biomedpress




