Abstract
Introduction: Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer in the world. Abnormal methylation can be one reason for this cancer. This study aimed to investigate the DNA promotor methylation status of cancer-associated genes (ATM, APC, CDO1, RB1, TP53, and WIF1) in patients with HNSCC.
Methods: Bisulfite conversion and methylation-sensitive high-resolution melting were used to analyze the DNA methylation levels in normal and tumor tissues in 44 patients.
Results: Significant differences in DNA methylation were observed between tumor and normal tissues for CDO1 and WIF1 genes in all subjects and subgroups (p < 0.05). In the T3 subgroup, a significant correlation was found between CDO1 gene methylation and age in normal tissue. The same correlation was detected for the WIF1 gene methylation in tumor tissue samples in the subgroup with T3 and normal tissue samples in the subgroup with T4 (p < 0.05). In all genes, no significant differences were found between the patient subgroups (T2, T3, T4 stage, primary/recurrent lesion, non-keratinizing/keratinizing SCC, age before/after 50, and smokers/non-smokers).
Conclusion: Changes in the expressions of CDO1 and WIF1 genes can affect the mechanisms of the occurrence and development of HNSCC. Methylation in the ATM, APC, RB1, and TP53 genes is not specific to HNSCC.
Introduction
Epigenetic gene regulation changes (such as DNA methylation, histone modification, chromatin remodeling, and non-coding RNA) and DNA mutation are the basis of the molecular mechanisms of cancerogenesis and tumorigenesis. Therefore, studying epigenetic markers for diagnosis, prognosis, and cancer prevention is particularly interesting. Detection of DNA methylation levels has recently been used as biomarkers for early diagnosis and prognosis to identify target genes for drug therapy.
The hypomethylation of single CpG dinucleotides has been one of the first changes found in tumor cells1. A large part of the investigation has been carried out during DNA methylation in the promoter region of various genes2, 3. There are changes in the methylation levels of general and tissue-specific cell genes in the ontogenesis in utero and post-natal periods.
The non-mutagenic etiology of cancer, such as disturbance of DNA methylation, is influenced by age, diet, transplacental, environmental, and occupational exposure. Moreover, its dual ability to be reversible and transgenerational demands a specific approach to interpreting results. Age-related DNA methylation is associated with different diseases4 and sex hormone status in some cancer types5. There is a significant gap in understanding the association between the age-expected DNA methylation level and cancer6. In particular, no data exist regarding pre- and postmenopausal differences in DNA methylation levels and their effects on cancer development.
The most intensive processes are in the genome of active proliferating cells, such as epithelial cells. Therefore, epithelial cells in various cancers are convenient models for studying epigenetic malformation. Head and neck squamous cell carcinoma (HNSCC) is an example of oncopathology with epithelial tissue damage.
HNSCC corresponds to the majority of cases (~90%) of head and neck cancers (HNCs)7. HNCs are rated 6–7 in global prevalence among cancers, with 700,000 newly registered cases and 470,000 deaths a year8. The therapeutic options for HNCs include surgical treatment, radiotherapy, and chemotherapy. The management tactics should be personalized by a multidisciplinary board comprising an oncological surgeon, a chemotherapist, a radiotherapist, a psychologist, a rehabilitation specialist, and a dentist9, 10.
Despite the advanced diagnostics, cancer screening, and HPV vaccination programs, 60–70% of the cases are diagnosed as late as during stage III–IV, associated with low life expectancy and high risks of recurrence11. Therefore, finding new potential targets for therapy is crucial.
The main risk factor for HNSCC is smoking. In some publications, it has been shown that smokers have a 10-fold higher risk of developing the disease than non-smokers. Smoking, in combination with frequent alcohol consumption, increases the risk more than 35-fold12, 13, 14. Other risk factors include ultraviolet and ionizing radiation, various toxic compounds, a weakened immune system, a diet low in vitamins A and B, age over 40, male, and some viral infections15.
DNA methylation also varies due to several factors, including age and disease status. Silenced genes are often methylated, while active genes remain largely unmethylated16, 17. DNA methylation is the most studied epigenetic mechanism in HNSCC. It allows us to observe changes in methylation patterns in a whole genome and individual genes that can be used to identify new disease biomarkers18. Various biomaterials can be used for studies, including liquid biopsy samples, which provide a noninvasive alternative for early cancer detection. In particular, Zhou C. studied 27 aberrantly methylated genes with altered expression and showed that FAM135B methylation is a favorable independent prognostic marker for overall survival in patients with HNSCC19.
We previously investigated the DNA methylation status of some tumor-associated genes (CDO1, MEST, RASSF1A, RASSF2, RASSF5, and WIF1) in patients with HNSCC20. We observed significant differences in DNA methylation levels between tumor and normal tissues in the CDO1 and WIF1 genes in all patient groups and subgroups (larynx and other cancers, SCC keratinizing and non-keratinizing, primary and recurrent tumor, and smokers and non-smokers)21. The methylation level in the CDO1 gene in tumor tissue was significantly increased in the T4 and T3 stage subgroups over the T2 stage22.
For further research, we investigated another group of tumors and normal tissue from patients with HNSCC. We chose other cancer-associated genes: ATM, APC, RB1, and TP53. We also used these earlier studied genes (CDO1 and WIF1), as they had shown notable results in other patients (Table 1). The selected genes play a considerable role in the regulation of cell proliferation, differentiation, and apoptosis, the disruption of which can lead to oncopathology.
This study aimed to investigate the DNA promotor methylation status of cancer-associated genes (ATM, APC, CDO1, RB1, TP53, and WIF1) in patients with HNSCC.
| Symbol | Gene name | Location | Exon count | Gene ID | Transcripts | MIM | Gene type | Gene function | References |
|---|---|---|---|---|---|---|---|---|---|
| APC GS; DP2; DP3; BTPS2; DESMD; DP2.5; PPP1R46 | Adenomatous Polyposis Coli | 5q22.2 | 20 | 5624 | NM_000038.6 | 611731 | protein-coding | tumor suppressor | 37; 39 |
| ATM AT1; ATA; ATC; ATD; ATE; ATDC; TEL1; TELO1 | Ataxia-telangiectasia-mutated | 11q22.3 | 67 | 472 | NM_000051.4 | 607585 | protein-coding | cell cycle regulation. | 42-44 |
| CDO1 (CDO-I) | Cysteine dioxygenase type 1 | 5q22.3 | 9 | 1036 | NM_001323565.2 | 603943 | protein-coding | tumor suppressor | 26-28 |
| TP53 (P53; BCC7; LFS1; BMFS5; TRP53) | Tumor Protein P53 | 17p13.1 | 11 | 7157 | NM_000546.6. | 191170 | protein-coding | tumor suppressor | 45-50 |
| RB1 (RB; pRb; OSRC; pp110; p105-Rb; PPP1R130; p110-RB1) | Retinoblastoma 1 | 13q14.2 | 27 | 5925 | NM_000321.3... | 614041 | protein-coding | cell cycle regulator, tumor suppressor | 51-53 |
| WIF1 (WIF-1) | WNT inhibitory factor 1 | 12q14.3 | 10 | 11197 | NM_007191.5 | 605186 | protein-coding | tumor suppressor | 19; 30-36 |
| Patient ID | Tumor origin | ICD-10 | TNM classification | Type of lesion | Histological glade | Gender | Age | Smoker |
|---|---|---|---|---|---|---|---|---|
| 1 | Tongue n=9 | C02.0 | T1N0M0 | Prim | SCC non-keratinizing | М | 50 | y |
| 2 | C02.1 | T2N0M0 | Prim | SCC non-keratinizing | F | 59 | n | |
| 3 | C02.1 | T3N0M0 | Rec | SCC keratinizing | F | 66 | y | |
| 4 | C02.1 | T3N0M0 | Rec | SCC non-keratinizing | F | 47 | n | |
| 5 | C02.1 | T3N0M0 | Prim | SCC keratinizing | М | 63 | n | |
| 6 | C02.1 | T3N0M0 | Prim | SCC keratinizing | М | 40 | n | |
| 7 | C02.1 | T3N0M0 | Prim | SCC keratinizing | M | 46 | n | |
| 8 | C02.1 | T3N1M0 | Prim | SCC non-keratinizing | М | 52 | n | |
| 9 | C02.1 | T4N2M0 | Prim | SCC keratinizing | М | 36 | n | |
| 10 | Oral cavity n=6 | C03.0 | T2N0M0 | Prim | SCC non-keratinizing | F | 59 | n |
| 11 | C03.0 | T3N0M0 | Prim | SCC keratinizing | M | 69 | y | |
| 12 | C03.0 | T4N0M0 | Prim | SCC keratinizing | М | 37 | y | |
| 13 | C03.1 | T4N0M0 | Prim | SCC non-keratinizing | М | 30 | n | |
| 14 | C03.1 | T4N0M0 | Prim | SCC non-keratinizing | F | 63 | n | |
| 15 | C03.1 | T4N0M0 | Prim | SCC non-keratinizing | F | 48 | n | |
| 16 | Floor of mouth n=3 | C04.1 | T2N0M0 | Prim | SCC keratinizing | F | 67 | y |
| 17 | C04.1 | T3N1M0 | Prim | SCC non-keratinizing | F | 66 | y | |
| 18 | C04.1 | T4N0M0 | Prim | SCC keratinizing | F | 64 | y | |
| 19 | Maxillary sinus n=6 | C31.0 | T3N0M0 | Prim | SCC non-keratinizing | M | 58 | y |
| 20 | C31.1 | T3N0M0 | Prim | SCC non-keratinizing | M | 55 | y | |
| 21 | C31.8 | T3N0M0 | Rec | SCC non-keratinizing | M | 64 | y | |
| 22 | C31.0 | T4N0M0 | Rec | SCC non-keratinizing | M | 41 | n | |
| 23 | С31.8 | T4aN0M0 | Prim | SCC non-keratinizing | M | 61 | n | |
| 24 | C31.0 | T4aN0M0 | Prim | SCC non-keratinizing | M | 67 | n | |
| 25 | Larynx n=20 | C32.1 | T1N2M0 | Prim | SCC non-keratinizing | М | 48 | n |
| 26 | C32.0 | T2N0M0 | Prim | SCC non-keratinizing | F | 55 | n | |
| 27 | С32.8 | T2N0M0 | Prim | SCC non-keratinizing | M | 70 | n | |
| 28 | С32.0 | T2N0M0 | Prim | SCC non-keratinizing | M | 69 | y | |
| 29 | C32.0 | T2N0M0 | Prim | SCC non-keratinizing | М | 58 | y | |
| 30 | C32.0 | T3N0M0 | Prim | SCC keratinizing | M | 64 | y | |
| 31 | C32.0 | T3N0M0 | Prim | SCC non-keratinizing | M | 62 | n | |
| 32 | C32.0 | T3N0M0 | Prim | SCC keratinizing | М | 62 | n | |
| 33 | C32.8 | T3N0M0 | Rec | SCC non-keratinizing | М | 64 | y | |
| 34 | C32.8 | T3N0M0 | Prim | SCC non-keratinizing | М | 69 | y | |
| 35 | C32.8 | T3N0M0 | Prim | SCC non-keratinizing | М | 58 | y | |
| 36 | C32.9 | T3N0M0 | Prim | SCC keratinizing | M | 61 | n | |
| 37 | C32.8 | T3N1M0 | Prim | SCC non-keratinizing | M | 49 | y | |
| 38 | C32.8 | T3N0M0 | Rec | SCC non-keratinizing | М | 69 | n | |
| 39 | C32.0 | T4N2M0 | Prim | SCC non-keratinizing | М | 64 | n | |
| 40 | C32.0 | T4N0M0 | Prim | SCC keratinizing | М | 71 | y | |
| 41 | C32.8 | T4aN0M0 | Prim | SCC keratinizing | M | 72 | y | |
| 42 | C32.8 | T4aN0M0 | Prim | SCC non-keratinizing | M | 58 | n | |
| 43 | С32.8 | T4aN2aM0 | Prim | SCC non-keratinizing | M | 70 | y | |
| 44 | C32.8 | T4aN2bM0 | Rec | SCC non-keratinizing | F | 81 | n |
| Genes | Forward primer sequence (5’ → 3’) | Reverse primer sequence (5’ → 3’) | Product size (bp*) |
|---|---|---|---|
| ATM | GTTGGTTATTGGTGGATATGG | TAATTCCAAAACCCAAACTCTTAAC | 696 |
| APC | GTTGGTTATTGGTGGATATGG | AACCTACAAAACCAAAAACCAACTA | 600 |
| CDO1 | GGGAGGATGA ATTTTATAGATTTG | TAAACTTCCATA ATAACCTACACCTC | 396 |
| RB1 | GATAGGGATGAGGTTTATAGTTATTTATTA | AAAATCCTATCACCATTCTACAAAC | 770 |
| TP53 | GGATTATTTGTTTTTATTTGTTATGG | CAAAACTCCACTCCTCTACCTAAAC | 495 |
| WIF1 | GAGTGATGTT TTAGGGGT | CCTCAACCA AAACTATTCC | 464 |
| Patients (n) Genes | Sample | All (n=44) | TNM ** | Type of lesion | SCC | Age | Smokers | ||||||
| T2 (n=7) | T3 (n=20) | T4 (n=15) | Prim (n=37) | Rec (n=7) | non-keratinizing (n=30) | keratinizing (n=14) | before 50 years (n=10) | (n=34) after 50 years | Yes (n=20) | No (n=24) | |||
| APC | T | 0.36±0.17 (0.01÷0.54) | 0.31±0.14 (0.05÷0.47) | 0.32±0.18 (0.01÷0.54) | 0.31±0.16 (0.01÷0.52) | 0.30±0.18 (0.01÷0.54) | 0.36±0.16 (0.01÷0.47) | 0.36±0.14 (0.01÷0.54) | 0.24±0.20 (0.01÷0.52) | 0.32±0.17 (0.01÷0.50) | 0.31±0.17 (0.01÷0.54) | 0.31±0.16 (0.01÷0.50) | 0.31±0.20 (0.01÷0.54) |
| N | 0.28±0.18 (0.01÷0.56) | 0.22±0.18 (0.07÷0.48) | 0.31±0.18 (0.04÷0.56) | 0.24±0.18 (0.01÷0.56) | 0.27±0.19 (0.01÷0.56) | 0.34±0.14 (0.01÷0.46) | 0.32±0.17 (0.01÷0.56) | 0.21±0.19 (0.01÷0.56) | 0.27±0.18 (0.04÷0.50) | 0.28±0.19 (0.01÷0.56) | 0.27±0.17 (0.01÷0.50) | 0.29±0.20 (0.01÷0.56) | |
| ATM | T | 0.35±0.18 (0.01÷0.54) | 0.31±0.20 (0.04÷0.54) | 0.34±0.19 (0.01÷0.54) | 0.37±0.18 (0.06÷0.54) | 0.34±0.19 (0.01÷0.54) | 0.38±0.19 (0.01÷0.53) | 0.39±0.18 (0.01÷0.54) | 0.28±0.18 (0.01÷0.53) | 0.30±0.23 (0.01÷0.53) | 0.37±0.17 (0.04÷0.54) | 0.33±0.19 (0.01÷0.54) | 0.37±0.18 (0.04÷0.54) |
| N | 0.30±0.19 (0.01÷0.57) | 0.27±0.22 (0.06÷0.57) | 0.30±0.19 (0.01÷0.56) | 0.29±0.17 (0.07÷0.49) | 0.31±0.19 (0.02÷0.57) | 0.27±0.21 (0.01÷0.49) | 0.36±0.18 (0.01÷0.57) | 0.20±0.18 (0.01÷0.55) | 0.29±0.19 (0.02÷0.50) | 0.30±0.19 (0.01÷0.57) | 0.30±0.18 (0.02÷0.57) | 0.30±0.20 (0.01÷0.56) | |
| CDO1 | T | 0.41±0.16* (0.18÷0.76) | 0.30±0.11 (0.18÷0.48) | 0.44±0.16* (0.18÷0.76) | 0.42±0.16* (0.18÷0.71) | 0.38±0.15* (0.18÷0.71) | 0.51±0.18* (0.26÷0.76) | 0.43±0.15* (0.18÷0.71) | 0.36±0.16* (0.18÷0.66) | 0.38±0.15* (0.18÷0.66) | 0.41±0.16* (0.18÷0.76) | 0.44*±0.16 (0.18÷0.76) | 0.36±0.16 (0.18÷0.71) |
| N | 0.20±0.11 (0.01÷0.53) | 0.20±0.10 (0.08÷0.34) | 0.19±0.10 (0.01÷0.34) | 0.19±0.11 (0.01÷0.32) | 0.19±0.11 (0.01÷0.34) | 0.23±0.11 (0.01÷0.34) | 0.21±0.10 (0.01÷0.34) | 0.17±0.10 (0.01÷0.32) | 0.17±0.13 (0.01÷0.32) | 0.21±0.09 (0.01÷0.34) | 0.20±0.11 (0.01÷0.34) | 0.20±0.10 (0.01÷0.34) | |
| TP53 | T | 0.10±0.06 (0.01÷0.20) | 0.08±0.05 (0.01÷0.18) | 0.09±0.06 (0.01÷0.20) | 0.11±0.06 (0.01÷0.20) | 0.10±0.06 (0.01÷0.20) | 0.10±0.08 (0.01÷0.20) | 0.10±0.06 (0.01÷0.18) | 0.10±0.04 (0.01÷0.17) | 0.10±0.06 (0.01÷0.20) | 0.10±0.06 (0.01÷0.20) | 0.10±0.07 (0.01÷0.20) | 0.10±0.05 (0.01÷0.18) |
| N | 0.09±0.07 (0.01÷0.21) | 0.08±0.07 (0.01÷0.19) | 0.09±0.06 (0.01÷0.20) | 0.09±0.07 (0.01÷0.21) | 0.09±0.07 (0.01÷0.21) | 0.07±0.05 (0.01÷0.16) | 0.08±0.07 (0.01÷0.21) | 0.09±0.06 (0.01÷0.21) | 0.15±0.04 (0.09÷0.21) | 0.07±0.06 (0.01÷0.20) | 0.10±0.07 (0.01÷0.20) | 0.07±0.06 (0.01÷0.21) | |
| RB1 | T | 0.26±0.12 (0.01÷0.40) | 0.21±0.12 (0.01÷0.36) | 0.27±0.13 (0.01÷0.40) | 0.27±0.12 (0.01÷0.38) | 0.26±0.13 (0.01÷0.40) | 0.27±0.13 (0.01÷0.36) | 0.28±0.11 (0.01÷0.40) | 0.24±0.15 (0.01÷0.38) | 0.27±0.10 (0.06÷0.39) | 0.25±0.14 (0.01÷0.40) | 0.26±0.12 (0.01÷0.39) | 0.27±0.14 (0.01÷0.40) |
| N | 0.25±0.11 (0.01÷0.40) | 0.26±0.11 (0.01÷0.38) | 0.26±0.11 (0.01÷0.40) | 0.23±0.10 (0.01÷0.36) | 0.26±0.10 (0.01÷0.40) | 0.23±0.14 (0.01÷0.36) | 0.26±0.10 (0.01÷0.40) | 0.23±0.12 (0.01÷0.36) | 0.22±0.10 (0.01÷0.34) | 0.24±0.12 (0.01÷0.40) | 0.25±0.10 (0.01÷0.38) | 0.26±0.12 (0.01÷0.40 | |
| WIF1 | T | 0.30±0.16* (0.08÷0.58) | 0.33±0.15 (0.14÷0.58) | 0.30±0.17 (0.08÷0.58) | 0.29±0.15* (0.09÷0.58) | 0.29±0.15* (0.08÷0.58) | 0.34±0.16* (0.09÷0.56) | 0.26±0.13 (0.08÷0.58) | 0.37±0.18* (0.08÷0.59) | 0.23±0.13* (0.09÷0.55) | 0.32±0.15 (0.09÷0.58) | 0.29*±0.16 (0.08÷0.58) | 0.32±0.16 (0.08÷0.58) |
| N | 0.16±0.08 (0.01÷0.42) | 0.21±0.05 (0.15÷0.30) | 0.18±0.10 (0.01÷0.42) | 0.11±0.05 (0.01÷0.22) | 0.16±0.07 (0.01÷0.42) | 0.18±0.08 (0.08÷0.26) | 0.16±0.09 (0.01÷0.42) | 0.15±0.08 (0.01÷0.30) | 0.10±0.06 (0.01÷0.20) | 0.18±0.08 (0.01÷0.20) | 0.14±0.06 (0.01÷0.26) | 0.18±0.11 (0.01÷0.42) |
Methods
Patient characteristics
The study involved 44 patients (34 men and 10 women) with HNSCC treated at A. Tsyb Medical Radiology Center, Obninsk, and P.A. Herzen Cancer Research Institute, Moscow. The study was approved by the A. Tsyb MRC Ethics Committee (approval № 634-17.11.2021).
For the first step of therapy, all patients underwent surgery. The absence of other treatments before surgery was the main inclusion criterion.
The sampling time duration was 6 months. All patients were sampled before starting further therapies. The characteristics of the patients are listed in Table 2. In total, 37 patients were diagnosed with primary HNSCC, and tumor recurrence after therapy was observed in seven cases. Squamous cell carcinoma of the tongue was observed in nine cases, cancer of the oral cavity was observed in six, cancer of the floor of the mouth was observed in three, cancer of the maxillary sinus was observed in six, and larynx cancer was observed in 20 patients. The non-keratinizing tumors were observed in 28 patients, with the keratinizing type observed in 16 patients [21]. Of the 44 patients, 26 were smokers. The mean age of the men was 58 (30÷72), and that of the women was 58 (47÷81) years old. Distribution by age in all patients is shown in Figure 1: four patients were 30–40 years of age (9%), eight patients were 41–50 years (18%), nine patients were 52–59 years (20%), 20 patients were 61–70 years (44%), and four patients were 71–81 (9%). The patients are classified according to the tumor, node, metastasis (TNM) system22, in which T1 - 2 (5%), T2 - 7 (16%), T3 - 20 (45%), and T4 - 15 (34%).
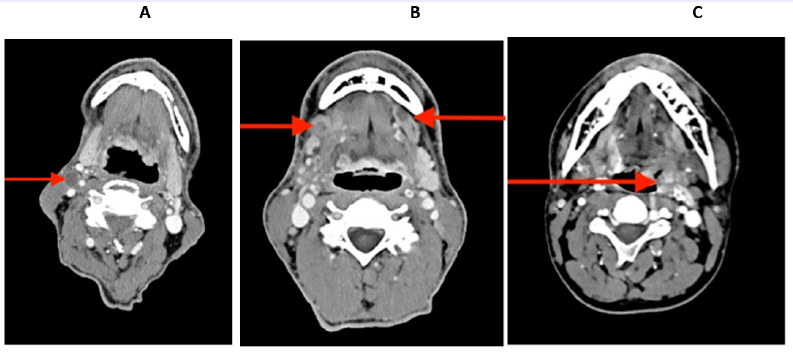
The pre-operative contrast-enhanced CT images of three patients with various diagnoses are shown in Figure 2.
Sampling
Tumor and normal tissue samples from each patient were obtained during surgery and stored at −20°C.
DNA extraction
DNA isolation from biomaterials was performed on microcolumns (K-SORB, № EX-514, Syntol, Russia) following the manufacturer's instructions.
DNA methylation analysis
Bisulfite conversion was performed using the EZ DNA Methylation-Lightning Kit (ThermoFisher EpiJET Bisulfite Conversion Kit, K1461, ThermoFisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions.
Methylation of the promoter regions of the genes was performed using the methylation-sensitive high-resolution melting (MS-HRM) method using the CFX 96 Connect real-time system (BioRad, USA).
The primers for the reaction were selected using Primer Blast software (Table 3). Ready-mix (PCR-Mix, M-428, Syntol, Russia) was used for two-step PCR. The program of amplification included 95°С — 5 min; (95°С — 15 s, 60°С — 30 s, 72°С — 45 s) × 30 cycles; (95°С — 15 s, 50°С — 30 s, 72°С — 45 s) × 25 cycles23. Furthermore, the intercalating dye EVA Green (Syntol, Russia) was added to the obtained products. Each sample was run in duplicate. The melting curve was constructed according to the following program: first stage - 95° - 30 s; second stage - 60° - 10 min; third stage — melting analysis in the range of 60–90° with a 0.2° step. MS-HRM was performed using Precision Melt Analysis Software, version 3 (BioRad, USA). A CFX96 amplifier (BioRad, USA) was used for PCR and MS-HRM. The methylation level was detected by fluorescence expressed in relative fluorescence units (RFU)24.
Statistical analysis
Statistical analysis of the data was carried out using R language. The method used was a chi-squared test. P values less than 0.05 were considered statistically significant.
Results
The average methylation level results of the DNA promoter for all normal and tumor tissues of the patients and subgroups are shown in Table 4. Significant differences in DNA methylation levels between the tumor and normal tissues were found for CDO1 and WIF1 genes in all patients (p < 0.05). Significant differences were also observed for CDO1 and WIF1 genes in the tumor and normal tissues in the different subgroups (p < 0.05). The exception was the CDO1 gene in a subgroup of patients with T2 and non-smokers. Regarding the WIF1 gene, the exceptions were found in subgroups of patients with T2 and T3, patients over 50, and non-smokers. The methylation levels of the DNA promoter of ATM, APC, RB1, and TP53 genes were insignificant. Furthermore, no significant differences were found between the subgroups (T2, T3, and T4 stage, with primary and recurrent lesions, non-keratinizing and keratinizing SCC, before and after 50 years of age, and smokers and non-smokers) of the patients.
Afterward, significant differences were observed in the methylation level between the tumor and normal tissues only for the CDO1 and WIF1 genes. Using different TNM systems, we analyzed the relationship between these data and age in the patient samples (Figure 3). We observed a significant positive correlation in the methylation level in the CDO1 gene with age in normal tissue from patients with T3 (Age/Normal: y = −0.3102 + 0.0085*x; r = 0.66; p = 0.002; r2 = 0.43). The correlation between the methylation level in CDO1 with age in tumor tissue from patients with T3 was not significant but was different (Age/Tumor3: y = −0.0641 + 0.0086*x; r = 0.42; p = 0.07; r2 = 0.17). We observed a significant correlation between methylation level in the WIF1 gene and age in the tumor tissue samples from the subgroup with T3 (Age/Tumor3: y = −0.4654 + 0.013*x; r = 0.59; p = 0.006; r2 = 0.35) and normal tissue samples in the subgroup with T4 (Age/Normal: y = −0.0002 + 0.0019*x; r = 0.52; p = 0.05; r2 = 0.27).
Discussion
Epigenetic disorders in tumor tissue cells in various types of oncopathology are as important as mutations, according to abundant literature data from the last decade. Early-stage epigenetic alterations can be identified and exploited to diagnose tumors early and predict cancer prognosis25, and DNA methylation can be used as a molecular target for cancer treatment because it is reversible via pharmacological inhibition of DNA methyltransferase.
(CDO-I) Cysteine dioxygenase type 1 protein-coding gene and tumor suppressor. CDO1 expression may be silenced by promoter DNA methylation. CDO1 can augment the generation of reactive oxygen species to induce apoptosis. Many studies have shown increased promoter methylation and, consequently, suppression of the CDO1 gene in various tumor cells26. In primary breast cancer patients and those with prostate cancer, the methylation level of the CDO1 gene's promoter region correlates strongly with tumor development. It can be utilized as a reliable prognostic indicator27. High levels of promoter CDO1 methylation have been observed in Barrett esophageal adenocarcinoma and prostate cancer patients28, 29. Our findings align with the evidence from the literature because the overall group of patients with HNSCC and subgroups had considerably higher CDO1 gene promoter methylation levels in tumor tissue than in normal tissue. Moreover, significant differences were found between smokers and non-smokers.
(WIF1) WNT inhibitory factor 1 is a protein-coding gene that is a tumor suppressor gene. These genes are epigenetically silenced in various cancers. The protein encoded by this gene functions to inhibit WNT proteins, which are extracellular signaling molecules that play a role in embryonic development. Wnt signal transduction dysregulation can contribute to head and neck cancer. Inhibition of Wnt signaling induces apoptosis and inhibits tumor growth in many cancer types30. Mutations in the genes encoding proteins in the Wnt (Wingless/Int-1) pathway are rare in HNSCC. Thus, this pathway is not considered significant for the pathogenesis of head and neck carcinomas31.
Few studies exist on the methylation levels of the Wnt gene. Our data indicated no significant differences in normal and tumor tissues from patients in terms of the DNA methylation level of the Wnt gene.
Methylation of the WIF1 gene, one of the antagonists of this pathway, is often associated with the development of this pathology32. WIF1 has the highest methylation frequency in oral carcinomas. WIF1 is methylated in 18% of patients with oral squamous cell carcinoma33 and 35% of patients with tongue carcinoma34, and it is significantly methylated in nasopharyngeal tumors35. In addition, non-small cell lung cancer frequently exhibits hypermethylation of the WIF1 (Wnt inhibitory factor-1) promoter region, even though patient characteristics, such as age, sex, and smoking history, are unrelated to the methylation status36.
Our study showed substantial variations in the average WIF1 promoter methylation levels between tumor and normal tissues in the entire patient population and in the subgroups. There were also significant differences in this parameter in the tumor tissue in the subgroups of patients with keratinizing and non-keratinizing HNSCC.
It is well known that DNA methylation is associated with cancer and aging. Aberrant DNA methylation frequently occurs with aging and illnesses such as cancer. DNA methylation status can become disrupted according to age and a particular disease stage. For example, promoter-specific hypermethylation and concurrent gene silence are linked to various malignancies37. We found a significant correlation between methylation level for the CDO1 and WIF1 genes in the normal and tumor tissues and age only in the subgroups with T3 and T4. These data indicate some dependence of the methylation level on the patient's age. The individual epigenetic features of the patient’s genome likely play an essential role in the results obtained. Thus, the study should be repeated with a larger sample size, and/or patients should be stratified according to other criteria.
Adenomatous polyposis coli (APC) is a tumor suppressor gene that inhibits cell proliferation through the Wnt/β-catenin signaling pathway. APC interacts with DNA repair proteins, DNA replication proteins, tubulin, and other components. It is not expressed in some types of cancer, in particular prostate, breast, and colorectal cancer. Lack of expression is associated with reduced survival in cancer patients. Moreover, APC regulates the response to chemotherapy in cancer cells38. The Wnt/β-catenin signaling pathway is activated by APC mutations in colorectal, endometrial, and prostate malignancies39. Hypermethylation of this gene has been detected in oral squamous cell carcinoma samples in some studies but not in patients with head and neck cancers. Additionally, there are no differences in the methylation of the APC gene between HNSCC patients and healthy individuals. The APC promoter was methylated in 7% of DNA samples taken from the plasma of a population free of cancer. It was unrelated to several other putative risk factors, such as age, tobacco and alcohol use, family history of cancer, diet, and nutrition. Other research failed to find a connection between clinical traits and outcomes, such as survival and aberrant methylation40, 41. Our results showed no significant differences in DNA methylation for the APC gene between tumor and normal tissues.
The ataxia–telangiectasia mutation (ATM) gene product is a well-known tumor suppressor essential for maintaining genomic stability. The mutated form of the ATM gene is involved in cell cycle control, apoptosis, oxidative stress, and telomere maintenance, and its role as a risk factor for cancer development is well established. In addition, several studies have been conducted to evaluate the risk of ATM gene mutations for breast cancer. In addition, several studies have been conducted to evaluate the screening of ATM gene mutations for the occurrence of breast cancer42, 43. In HNSCC, the ATM gene promoter is a target for abnormal hypermethylation. Reduced ATM function through epigenetic silencing is a likely mechanism contributing to HNSCC and other tumor types, given its significant role in maintaining genome integrity and the causal role that genome instability plays in cancer onset and progression. It has been found that hypermethylation of the ATM promoter is significantly correlated with a decrease in the average survival rate of patients. Furthermore, this link is unrelated to other possible predictive elements, such as tumor size, lymph node condition, clinical stage, and history of tobacco and alcohol use. Thus, these data indicate that hypermethylation of the ATM promoter can be used as a prognostic factor in HNSCC44. However, our data revealed no significant differences in DNA methylation levels for the ATM gene in the tumor and normal tissues of patients.
Functional properties caused by tumor protein p53 (TP53) gene mutations are involved in cancer development and progression45, 46. Somatic mutation is one of the most prevalent changes in the TP53 gene in human tumors. Moreover, the underlying cause of Li-Fraumeni syndrome, which predisposes patients to various early-onset cancers, is germline mutation. TP53 gene alterations are also possible prognostic and predictive indicators and prospective drug targets47. According to a thorough integrative genomic study, most somatic genomic changes in HNSCC are mutations in the TP53 gene. Invasion, metastasis, genomic instability, and cancer cell proliferation are all facilitated by TP53 mutations, which can result in a loss of wild-type p53 activity or an increase in those activities. Interestingly, aggressiveness and worse survival following surgical treatment of HNSCC are related to disruptive TP53 mutations in tumor DNA48. Harris C.C. proposed that genetic analysis of the p53 gene in exfoliated cells detected in either body fluids or tissue biopsies might identify people at elevated cancer risk because mutations in the p53 gene can arise in precancerous lesions in the lung, breast, esophagus, and colon49. Hypermethylation of the promoter region of TP53 is often associated with a decrease in its activity and can even lead to its silence and, thus, to the loss of its function. This, in turn, contributes to malignant transformation50. Unfortunately, our research showed no significant differences in DNA methylation for the TP53 gene between tumor and normal tissues in the patients.
The tumor suppressor retinoblastoma gene (RB1) is essential for controlling the cell cycle by increasing G1/S arrest and limiting growth by blocking the E2F transcription factor. Even aneuploidy can result from RB1 loss, dramatically increasing the chance of developing cancer. The RB1 gene is part of a large gene family that includes RBL1 and RBL2, each encoding the structurally related proteins pRb, p107, and p13051. One frameshift and seven non-synonymous missense mutations were discovered in RB1 (pocket domain and spacer region) sequencing analysis. In HNC tumor samples, an RB1 promoter methylation study showed that 16% of its cytosines (3% in CpG) were methylated52. Sabir M. and colleagues indicated that genetic and epigenetic RB1 changes might contribute to the pathogenesis of HNSCC in the Pakistani population53. However, our study showed no significant differences in DNA methylation for the RB1 gene between tumor and normal tissues in our examined patients.
Even though in our patients with HNSCC, in the tumor-associated genes APC, ATM, TP53, and RB1, no significant changes were observed in the methylation level between normal and tumor tissue, these genes are not specific for epigenetic analysis.
We also did not find significant differences in any genes (T2, T3, and T4 stages, with primary and recurrent lesion, non-keratinizing and keratinizing SCC, age before and after 50 years, and smokers, and non-smokers) of the patients. This may be due to the small number of patients examined.
Conclusions
Thus, we can conclude that changes in the expressions of the CDO1 and WIF1 genes can affect the occurrence and development of HNSCC and can be considered prognostic and diagnostic markers for this pathology. Future application of molecular–genetic and epigenetic studies on HNSCC can serve as the basis for target therapy and contribute to personalized medicine.
Abbreviations
None.
Acknowledgments
None.
Author’s contributions
Kurevlev S., molecular-genetic investigation, development of research design;
Funding
The study was done with the financial support of the Russian Federation represented by the Ministry of Education and Science of Russia; Agreement dated October 7, 2021 No. 075-15-2021-1356 (internal number of the Agreement 15.SIN.21.0011); (ID: RF 0951.61321X0012).
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study protocol was approved by the Biomedical Ethics Committee of the A.F. Tsyb Medical Radiology Research Center Branch of the Federal State Institution "National Medical Research Center of Radiology" of the Ministry of Health of the Russian Federation (protocol № 634 from 17.11.2021).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Esteller
M.,
Corn
P.G.,
Baylin
S.B.,
Herman
J.G.,
A gene hypermethylation profile of human cancer. Cancer research.
2001;
61
(8)
:
3225-9
.
-
Gupta
Sh.,
Kumar
P.,
Maini
J.,
Kaur
H.,
Das
B.C.,
Epigenetic biomarkers in head and neck cancer. Journal of Cancer Genetics and Biomarkers.
2018;
1
(2)
:
41-50
.
View Article Google Scholar -
Castilho
R.M.,
Squarize
C.H.,
Almeida
L.O.,
Epigenetic modifications and head and neck cancer: implications for tumor progression and resistance to therapy. International Journal of Molecular Sciences.
2017;
18
(7)
:
1506
.
View Article PubMed Google Scholar -
Yasmeen
S.,
Bejaoui
Y.,
El Hajj
N.,
DNA Methylation Biomarkers in Aging and Age-Related Diseases. Review. Front. Genet., Sec. Frontiers in Genetics.
2020;
11
:
171
.
View Article Google Scholar -
Johansson
A.,
Palli
D.,
Masala
G.,
Grioni
S.,
Agnoli
C.,
Tumino
R.,
Epigenome-wide association study for lifetime estrogen exposure identifies an epigenetic signature associated with breast cancer risk. Clinical Epigenetics.
2019;
11
(1)
:
66
.
View Article PubMed Google Scholar -
Chen
L.,
Ganz
P.A.,
Sehl
M.E.,
DNA Methylation, Aging, and Cancer Risk: A Mini-ReviewFront. Bioinform., Sec. Genomic Analysis 2022.
View Article Google Scholar -
Rahman
Q.B.,
Iocca
O.,
Kufta
K.,
Shanti
R.M.,
Global Burden of Head and Neck Cancer. Oral and Maxillofacial Surgery Clinics of North America.
2020;
32
(3)
:
367-75
.
View Article PubMed Google Scholar -
Siegel
R.L.,
Miller
K.D.,
Fuchs
H.E.,
Jemal
A.,
Cancer statistics, 2022. CA: a Cancer Journal for Clinicians.
2022;
72
(1)
:
7-33
.
View Article PubMed Google Scholar -
Hashim
D.,
Genden
E.,
Posner
M.,
Hashibe
M.,
Boffetta
P.,
Head and neck cancer prevention: from primary prevention to impact of clinicians on reducing burden. Annals of Oncology : Official Journal of the European Society for Medical Oncology.
2019;
30
(5)
:
744-56
.
View Article PubMed Google Scholar -
Johnson
D.E.,
Burtness
B.,
Leemans
C.R.,
Lui
V.W.,
Bauman
J.E.,
Grandis
J.R.,
Head and neck squamous cell carcinoma. Nature Reviews. Disease Primers.
2020;
6
(1)
:
92
.
View Article PubMed Google Scholar -
Pfister
D.G.,
Spencer
S.,
Adelstein
D.,
Adkins
D.,
Anzai
Y.,
Brizel
D.M.,
Head and Neck Cancers, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network : JNCCN.
2020;
18
(7)
:
873-98
.
View Article PubMed Google Scholar -
Marcu
L.G.,
Yeoh
E.,
A review of risk factors and genetic alterations in head and neck carcinogenesis and implications for current and future approaches to treatment. Journal of Cancer Research and Clinical Oncology.
2009;
135
(10)
:
1303-14
.
View Article PubMed Google Scholar -
WHO.int [Electronic resourse]: The world health report 2002: Reducing Risks, Promoting Healthy Life. – Available from: http://www.who.int/ whr/2002/en/whr02_en.pdf?ua=1. – Date of access: 20.06.2018. ISBN: 9241562072.
.
-
WHO.int [Electronic resourse]: WHO global report on trends in tobacco smoking 2000–2025, 2015. – Available from: apps.who.int/iris/bitstre am/10665/156262/1/9789241564922_eng. – Date of access: 20.06.2018..
.
-
Global Cancer Observatory. International Agency for Research on Cancer. World Health Organization. Available at: https://gco.iarc.fr/. Accessed on June 06, (2021)..
.
-
Demokan
S.,
Dalay
N.,
Role of DNA methylation in head and neck cancer. Clinical Epigenetics.
2011;
2
(2)
:
123-50
.
View Article PubMed Google Scholar -
Maegawa
S.,
Lu
Y.,
Tahara
T.,
Lee
J.T.,
Madzo
J.,
Liang
S.,
Caloric restriction delays age-related methylation drift. Nature Communications.
2017;
8
(1)
:
539
.
View Article PubMed Google Scholar -
Ovchinnikov
D.A.,
Cooper
M.A.,
Pandit
P.,
Coman
W.B.,
Cooper-White
J.J.,
Keith
P.,
Tumor-suppressor gene promoter hypermethylation in saliva of head and neck cancer patients. Translational Oncology.
2012;
5
(5)
:
321-6
.
View Article PubMed Google Scholar -
Zhou
C.,
Ye
M.,
Ni
S.,
Li
Q.,
Ye
D.,
Li
J.,
Shen
Z.,
Deng
H.,
DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics.
2018;
13
(4)
:
398-409
.
View Article Google Scholar -
Kurevlev
S.V.,
Tskhovrebova
L.V.,
Aghajanyan
A.V.,
Fatkhudinov
T.K.,
Gordon
K.B.,
Azova
M.M.,
Methylation of the tumor associated genes in head and neck squamous cell carcinoma. Head and Neck Tumors.
2022;
12
(4)
:
61-70
.
View Article Google Scholar -
Mete
O.,
Wenig
B.M.,
Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Overview of the 2022 WHO Classification of Head and Neck Neuroendocrine Neoplasms. Head Neck Pathol. (2022) 16(1):123-142.
.
View Article Google Scholar -
Amin
Mahul B,
Edge
Stephen B,
Greene
Frederick L,
Byrd
David R,
Brookland
Robert K,
Washington
Mary Kay,
Gershenwald
Jeffrey E,
Compton
Carolyn C,
Hess
Kenneth R,
Sullivan
Daniel C,
AJCC cancer staging manualSpringer 2017.
Google Scholar -
Krasnyi
A.M.,
Kurevlev
S.V.,
Sadekova
A.A.,
Sefihanov
T.G.,
Kometova
V.V.,
Rodionov
V.V.,
Primary tumor gene methylation profile in patients with luminal HER2-negative breast cancer when metastasizing to regional lymph nodes (Russian). Biomedicinskaya himia..
2021;
67
(1)
:
88-94
.
-
S\lomka
M.,
Sobalska-Kwapis
M.,
Wachulec
M.,
Bartosz
G.,
Strapagiel
D.,
High Resolution Melting (HRM) for High-Throughput Genotyping-Limitations and Caveats in Practical Case Studies. International Journal of Molecular Sciences.
2017;
18
(11)
:
2316
.
View Article PubMed Google Scholar -
Yamashita
K.,
Hosoda
K.,
Nishizawa
N.,
Katoh
H.,
Watanabe
M.,
Epigenetic biomarkers of promoter DNA methylation in the new era of cancer treatment. Cancer Science.
2018;
109
(12)
:
3695-706
.
View Article PubMed Google Scholar -
Brait
M.,
Ling
S.,
Nagpal
J.K.,
Chang
X.,
Park
H.L.,
Lee
J.,
Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS One.
2012;
7
(9)
:
e44951
.
View Article PubMed Google Scholar -
Minatani
N.,
Waraya
M.,
Yamashita
K.,
Kikuchi
M.,
Ushiku
H.,
Kojo
K.,
Prognostic significance of promoter DNA hypermethylation of cysteine dioxygenase 1 (CDO1) gene in primary breast cancer. PLoS One.
2016;
11
(1)
:
e0144862
.
View Article PubMed Google Scholar -
Meller
S.,
Zipfel
L.,
Gevensleben
H.,
Dietrich
J.,
Ellinger
J.,
Majores
M.,
CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics.
2016;
11
(12)
:
871-80
.
View Article PubMed Google Scholar -
Kojima
K.,
Yamashita
K.,
Ushiku
H.,
Katoh
H.,
Ishii
S.,
Tanaka
T.,
The clinical significance of cysteine dioxygenase type 1 methylation in Barrett esophagus adenocarcinoma. Diseases of the Esophagus.
2017;
30
(3)
:
1-9
.
View Article PubMed Google Scholar -
Yang
T.M.,
Leu
S.W.,
Li
J.M.,
Hung
M.S.,
Lin
C.H.,
Lin
Y.C.,
WIF-1 promoter region hypermethylation as an adjuvant diagnostic marker for non-small cell lung cancer-related malignant pleural effusions. Journal of Cancer Research and Clinical Oncology.
2009;
135
(7)
:
919-24
.
View Article PubMed Google Scholar -
He
B.,
You
L.,
Uematsu
K.,
Xu
Z.,
Lee
A.Y.,
Matsangou
M.,
A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia (New York, N.Y.).
2004;
6
(1)
:
7-14
.
View Article PubMed Google Scholar -
Yeh
K.T.,
Chang
J.G.,
Lin
T.H.,
Wang
Y.F.,
Chang
J.Y.,
Shih
M.C.,
Correlation between protein expression and epigenetic and mutation changes of Wnt pathway-related genes in oral cancer. International Journal of Oncology.
2003;
23
(4)
:
1001-7
.
View Article PubMed Google Scholar -
Paluszczak
J.,
Sarbak
J.,
Kostrzewska-Poczekaj
M.,
Kiwerska
K.,
Jarmuż-Szymczak
M.,
Grenman
R.,
The negative regulators of Wnt pathway-DACH1, DKK1, and WIF1 are methylated in oral and oropharyngeal cancer and WIF1 methylation predicts shorter survival. Tumour Biology.
2015;
36
(4)
:
2855-61
.
View Article PubMed Google Scholar -
Pannone
G.,
Bufo
P.,
Santoro
A.,
Franco
R.,
Aquino
G.,
Longo
F.,
WNT pathway in oral cancer: epigenetic inactivation of WNT-inhibitors. Oncology Reports.
2010;
24
(4)
:
1035-41
.
PubMed Google Scholar -
Supic
G.,
Kozomara
R.,
Jovic
N.,
Zeljic
K.,
Magic
Z.,
Hypermethylation of RUNX3 but not WIF1 gene and its association with stage and nodal status of tongue cancers. Oral Diseases.
2011;
17
(8)
:
794-800
.
View Article PubMed Google Scholar -
Chan
S.L.,
Cui
Y.,
van Hasselt
A.,
Li
H.,
Srivastava
G.,
Jin
H.,
The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Laboratory Investigation.
2007;
87
(7)
:
644-50
.
View Article PubMed Google Scholar -
Morgan
A.E.,
Davies
T.J.,
Mc Auley
M.T.,
The role of DNA methylation in ageing and cancer. The Proceedings of the Nutrition Society.
2018;
77
(4)
:
412-22
.
View Article PubMed Google Scholar -
Stefanski
C.D.,
Prosperi
J.R.,
Wnt-Independent and Wnt-Dependent Effects of APC Loss on the Chemotherapeutic Response. International Journal of Molecular Sciences.
2020;
21
(21)
:
7844
.
View Article PubMed Google Scholar -
Katoh
M.,
Katoh
M.,
Molecular genetics and targeted therapy of WNT-related human diseases (Review). International Journal of Molecular Medicine.
2017;
40
(3)
:
587-606
.
View Article PubMed Google Scholar -
Strzelczyk
J.K.,
Krakowczyk
\L.,
Owczarek
A.J.,
Aberrant DNA methylation of the p16, APC, MGMT, TIMP3 and CDH1 gene promoters in tumours and the surgical margins of patients with oral cavity cancer. Journal of Cancer.
2018;
9
(11)
:
1896-904
.
View Article PubMed Google Scholar -
Ali
J.,
Sabiha
B.,
Jan
H.U.,
Haider
S.A.,
Khan
A.A.,
Ali
S.S.,
Genetic etiology of oral cancer. Oral Oncology.
2017;
70
:
23-8
.
View Article PubMed Google Scholar -
Stucci
L.S.,
Internò
V.,
Tucci
M.,
Perrone
M.,
Mannavola
F.,
Palmirotta
R.,
The ATM Gene in Breast Cancer: Its Relevance in Clinical Practice. Genes.
2021;
12
(5)
:
727
.
View Article PubMed Google Scholar -
Marabelli
M.,
Cheng
S.C.,
Parmigiani
G.,
Penetrance of ATM Gene Mutations in Breast Cancer: A Meta-Analysis of Different Measures of Risk. Genetic Epidemiology.
2016;
40
(5)
:
425-31
.
View Article PubMed Google Scholar -
Ai
L.,
Vo
Q.N.,
Zuo
C.,
Li
L.,
Ling
W.,
Suen
J.Y.,
Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiology, Biomarkers & Prevention.
2004;
13
(1)
:
150-6
.
View Article PubMed Google Scholar -
Lindemann
A.,
Takahashi
H.,
Patel
A.A.,
Osman
A.A.,
Myers
J.N.,
Targeting the DNA Damage Response in OSCC with TP53 Mutations. Journal of dental research.
2018;
97
(6)
:
635-44
.
View Article Google Scholar -
Sasaki
K.,
Takahashi
S.,
Ouchi
K.,
Otsuki
Y.,
Wakayama
S.,
Ishioka
C.,
Different impacts of TP53 mutations on cell cycle-related gene expression among cancer types. Scientific Reports.
2023;
13
(1)
:
4868
.
View Article PubMed Google Scholar -
Olivier
M.,
Hollstein
M.,
Hainaut
P.,
TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor Perspectives in Biology.
2010;
2
(1)
:
a001008
.
View Article PubMed Google Scholar -
Nathan
C.A.,
Khandelwal
A.R.,
Wolf
G.T.,
Rodrigo
J.P.,
Mäkitie
A.A.,
Saba
N.F.,
TP53 mutations in head and neck cancer. Molecular Carcinogenesis.
2022;
61
(4)
:
385-91
.
View Article PubMed Google Scholar -
Harris
C.C.,
p53 tumor suppressor gene: at the crossroads of molecular carcinogenesis, molecular epidemiology, and cancer risk assessment. Environmental health perspectives.
1996;
104
(suppl 3)
:
435-9
.
View Article Google Scholar -
Golusinski
P.,
Lamperska
K.,
Pazdrowski
J.,
Golusinski
W.,
Analiza wystepowania mutacji w obrebie genu TP53 u chorych na raka płaskonabłonkowego głowy i szyi [Analysis of mutations within the TP53 gene in patients with squamous cell carcinoma of the head and neck]. Otolaryngol Pol.
2011;
65
(2)
:
114-21
.
View Article Google Scholar -
Di Fiore
R.,
D'Anneo
A.,
Tesoriere
G.,
Vento
R.,
RB1 in cancer: different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. Journal of Cellular Physiology.
2013;
228
(8)
:
1676-87
.
View Article PubMed Google Scholar -
Rusin
P.,
Markiewicz
L.,
Majsterek
I.,
Uwarunkowania genetyczne nowotworów głowy i szyi [Genetic predeterminations of head and neck cancer]. Postepy Hig Med Dosw.
2008;
(62)
:
490-501
.
-
Sabir
M.,
Baig
R.M.,
Ali
K.,
Mahjabeen
I.,
Saeed
M.,
Kayani
M.A.,
Retinoblastoma (RB1) pocket domain mutations and promoter hyper-methylation in head and neck cancer. Cellular Oncology (Dordrecht).
2014;
37
(3)
:
203-13
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 11 No 1 (2024)
Page No.: 6164-6178
Published on: 2024-01-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 3215 times
- PDF downloaded - 1093 times
- XML downloaded - 118 times
 Biomedpress
Biomedpress




