Abstract
Background: Chronic kidney disease (CKD) is an ailment marked by a reduction in the glomerular filtration rate or the presence of proteinuria. Millions of people are affected worldwide, and so far, the mechanisms underlying these effects remain mostly unknown. Recently, the role of microRNAs (miRNAs) in several cellular processes associated with the development of diseases has been discovered. Studies have suggested that miR-149-5p may play a role in renal function and kidney disorders and be linked with inflammation, fibrosis, or apoptosis within the kidneys. In addition, miR-149-5p often targets signaling pathways, including TGF-β, Wnt/β-catenin, and NF κB. Hence, this study focused on miR-149-5p and its biomarker, interleukin-6 (IL-6), as potential indicators of CK D. Computational analyses were employed for miR-149-5p identification within CKD genome sequences, revealing its potential involvement in disease processes.
Methods: In this study, a total of 20 patient and normal blood samples were collected and stored for analysis. To detect miR-149-5p in CKD, target scanning, miRbase, and the National Centre for Biotechnology Information (NCBI) database were used, whereas RNA fold, melt curve, and Ct were utilized to construct the secondary structure and analyze miR-149-5p levels. A similar investigation was conducted to determine the IL-6 gene’s expression levels.
Results: Upon thoroughly studying the secondary structure, hsa-miR-149-5p’s lowest free energy was found to be −52.70 kCal. Additionally, dys-regulation of miR-149-5p and IL-6 expression in individuals with CKD was observed. miR-149-5p down-regulation and IL-6 over-expression pointed toward the potential role of these two molecules in the pathogenesis of CKD.
Conclusion: The computational techniques utilized explain miR-149-5p’s role as a diagnostic, predictive, and potentially effective therapeutic target for CKD. Moreover, these findings contribute to a better understanding of CKD, along with miR-149-5p’s role in developing novel treatments for this disease.
Introduction
CKD is an ailment marked by a reduction in glomerular filtration rate or the presence of proteinuria, with diabetic nephropathy and hypertension being the primary cause and renal damage being the main factor in most cases. Poor appetite, lethargy, insomnia, muscle cramps, itching, and swollen feet and ankles are the common symptoms of CKD. Two common indicators of CKD are elevated levels of creatinine and reduced albumin. Because a conclusive cure is not available for this condition, the main aim from a clinical perspective is to reduce the progression of renal failure, typically addressing sequelae such as left ventricular hypertrophy, vascular calcification, and anemia1, 2, 3, 4. Numerous studies have reported microRNAs (miRNAs) as being innovative tools for disease diagnosis or potential targets for therapy. miRNAs are non-coding RNA (ncRNA) molecules that mainly participate in the post-transcriptional regulation of genes and are widely studied in the context of CKD. The main sources of miRNAs are plasma, serum, urine, or exosomes, which are small membrane vesicles containing various proteins, lipids, and nucleic acids5. MiR-149-5p, a conserved mRNA, has been identified as a regulator of physiological processes such as inflammatory responses, adipogenesis, and cell proliferation. This molecule is expressed in diverse tissues, such as the brain, liver, and adipose tissue, and affects gene expression as well as cellular activities (differentiation, proliferation, and apoptosis). Recent studies indicate its function as an oncogene, along with its role in regulating pathways like the Wnt signaling pathway, crucial for cancer development and progression. Additionally, miR-149-5p plays a vital role in the immune response and inflammation regulation, whereas miR-149-5p dysregulation has been associated with numerous diseases like cancer, metabolic disorders, and cardiovascular diseases6, 7. miR-149-5p often targets signaling pathways, including TGF-β, Wnt/β-catenin, and NFκB8, 9, 10. As miRNAs can serve as biomarkers, changes in the expression levels of miR-149-5p may provide insights into disease progression and severity. Meanwhile, interleukin-6 (IL-6) plays a pivotal role in the pathogenesis of CKD, promoting inflammation, fibrosis, and cardiovascular problems11. Furthermore, miR-149-5p, when dysregulated, may influence IL-6 expression, potentially amplifying the inflammatory and fibrotic processes in CKD. Therefore, understanding the role of miR-149-5p in CKD could open avenues for therapeutic intervention. Modulating the expression of this miRNA might be explored as a potential strategy to mitigate CKD-associated pathologies.
Methods
Retrieval of CKD sequences and miRNAs
Data on human genome sequences was obtained from the National Center for Biotechnology Information (NCBI) web portal for the International Nucleotide Sequence Database Consortium. The CKD genome sequence was located using a free search engine using the search term “chronic kidney disease genome sequence in Homo sapiens.” Following the removal of repetitive and inadequate sequences, a local nucleotide database dedicated to CKD-specific genome sequences was established. The reference sequence was obtained from human-reported miRNAs (http://www.mirbase.org/). The aforementioned CKD database was then queried to identify homologs within the miRNA dataset12.
Identification of precursor miRNAs
Mature miRNAs were used as a starting point for searches to find homologs in the CKD database. miRNA sequences were used as search queries in homology searches using the Basic Local Alignment Search Tool (BLAST) 2.2.26+ against a recently created local database specific to hypothyroidism, employing 0.01 as the e-value threshold while keeping all other parameters at their default levels. For sequences with a maximum of three mismatches, BLAST was used to validate the non-protein encoding features. The aligned section was then designated as a potential precursor (pre)-miRNA sequence12.
Validation of candidate pre- miRNAs and their targets
Using RNA fold, a secondary structure was derived, yielding a mature miRNA sequence within the CKD sequence. Several criteria were employed for confirmation: 1) the RNA structure should exhibit a suitable stem-loop hairpin structure, 2) the mature miRNA should be located on one side of the hairpin structure, 3) the miRNA must have fewer than seven mismatches with the complementary miRNA in the opposing arm of the structure, and 4) the secondary structure should possess more negative energy and an A+U content of between 40 and 70%. Additionally, target prediction was performed using TargetScan to help find possible targets.
Sample collection
This study received approval from the Institutional Ethics Committee (581/03/2023/UG/S RB/SMCH), and all samples were gathered in strict adherence to the Helsinki Declaration. The sample size for the study was calculated using Gpower, and a set of 20 blood samples, comprising both CKD and normal cases, was acquired from patients who provided informed consent through the Department of Medicine at aveetha Medical College and Hospitals. The Department of Biochemistry at Saveetha Medical College and Hospitals confirmed the diagnosis of hypothyroidism. Following collection, the samples underwent centrifugation. While spinning the blood with an anticoagulant, the lighter plasma formed the upper layer and was carefully extracted13. This was then preserved in a deep freezer at −20 °C for subsequent analysis.
Inclusion and exclusion criteria
The study only accepted participants who were over 18 years old and able to provide informed consent. Individuals with secondary complications, such as hypertension or diabetes were excluded from participation.
RNA extraction and quantification
TRIzol reagent (Invitrogen, Carlsbad, CA, USA), which was added to the plasma, was used for total RNA extraction following the manufacturer's instructions. Using a NanoDrop 2000 Lite spectrophotometer from Thermo Fisher Scientific, both the quantity and quality of the extracted RNA were evaluated. After that, these samples were kept at −20 °C for later analysis14.
Reverse transcription
The obtained RNA was subjected to reverse transcription. This process includes combining the RNA sample with an oligo (dT)18 primer (Promega, 50 μM) for genes and a universal adapter for miRNAs, along with deoxyribonucleotide triphosphates (dNTPs, 10mM each New England Biolabs, Inc.) and nuclease-free water. This mixture was then incubated for 5 minutes at 65 °C and then immediately cooled, resulting in a volume of 10 μl. Subsequently, the mixture was supplemented with 5x prime buffer (New England Biolabs Inc.), murine RNase inhibitor (New England Biolabs, Inc.), reverse transcriptase (New England Biolabs, Inc.), and nuclease-free water, resulting in a final volume of 20 μl. This mixture was then subjected to incubation in a PCR (MiniAmp Plus thermal cycler, Thermo Fisher) under the following conditions: 10 minutes at 30 °C, 30 minutes at 42 °C, and 5 minutes at 95 °C, followed by a final incubation at 4 °C. cDNA was quantified using a Nanodrop Lite spectrophotometer and then stored at −20 °C for further studies14, 15.
Expression using qRT-PCR
For the miR-149-5p and IL-6 genes, expression studies were conducted using the cDNA produced using Sybr Green (Takara, Japan). GAPDH was used as the housekeeping control for IL-6, and U6 was used for miR-149-5p. The primers required for this process were purchased from Origene, and the BioRad CFX96 Realtime System was used for expression experiments. The temperatures for PCR cycling were as follows: initial denaturation for one cycle at 95 ºC for 30 seconds, denaturation for 5 seconds at 95 ºC, and annealing for 30 seconds for a maximum of 40 cycles with a melt curve. The 2^-∆∆Cq technique was used to calculate the gene expression for each test in duplicate 14,15.
Statistical analysis
The mean of the duplicate experiments with the standard error of the mean (SEM) was calculated. Student’s t-test was applied using GraphPad Prism 10.1.0. to assess differences between the groups, with significance considered at a value less than 0.05 (*). Meanwhile, Tukey’s test was used to determine the statistical significance between the groups.
| S. No | Structure | Sequence |
|---|---|---|
| 1 | Stem-loop | GCCGGCGCCCGAGCUCUGGCUCCGUGUCU UCACUCCCGUGCUUGUCCGAGGAGGGAGGGAGGG ACGGGGGCUGUGCUGGGGCAGCUGGA |
| 2 | Mature miRNA | UCUGGCUCCGUGUCUUCACUCCC |
| Source miRNA | Source organism | Pre-miRNA length | Minimum Free Energy | Mature Sequence | Match Extent | Strand | A+U% |
|---|---|---|---|---|---|---|---|
| miR-149-5p | Homo sapiens | 89 | - 52.70 kcal | UCUGGCUCCGUGU CUUCACUCCC | 23/23 | 5p | 26.9 |
| S. No | Target Protein | Molecular function | Molecular function |
|---|---|---|---|
| 1 | Interleukin 6 | Multifunctional cytokine | Cellular function |
| 2 | BCL2 binding component 3 | Binding of BCL2 | Cell apoptosis |
| 3 | Transmembrane protein 234 | Signal transmission | Transfer of molecules across the membrane |
| 4 | Tetraspanin 14 | Enzyme binding activity | Notch signalling |
| 5 | Zinc finger protein 74 | transcription | Inhibits acetylation |
Results
Identification of pre-miRNA and its secondary structure
MiRNAs are crucial in modulating gene expression, exerting influence over the development and progression of diseases; thus, identifying the specific miRNA associated with CKD holds promise for early detection and therapeutic interventions. Employing a computational approach, we embarked on the task of identifying these miRNAs. To accomplish this, precursor miRNAs were collected from the miRbase database, while the CKD sequences were sourced from the NCBI database. After the meticulous examination of a substantial number of sequences, a single miRNA, hsa-miR-149a-5p, was discerned within the CKD genome sequences. This discovery was facilitated by employing RNA fold, which revealed hsa-miR-149-5p’s mature sequence, with the lowest free energy of 52.80 kcal.
Figure 1 illustrates the secondary structure of hsa-miR-149-5p, while Table 1 provides the stem-loop and mature sequence of miR-149-5p. Additionally, Table 2 presents comprehensive information regarding the length of the pre-miRNA, minimum free energy, mature sequence, match extent, and A+U% content of hsa-miR-149-5p.
Identification of targets
TargetScan analysis pinpoints potential targets for the specific miRNA. Through this approach, several significant transcripts targeted by miR-149-5p, including IL-6, cyclin 1, BCL-2, cell division cycle 73, and transferrin receptor, among others, were identified. Table 3 provides an overview of these targets, along with their associated molecular functions and biological processes.
Gene expression analysis of miR-149-5p and IL-6
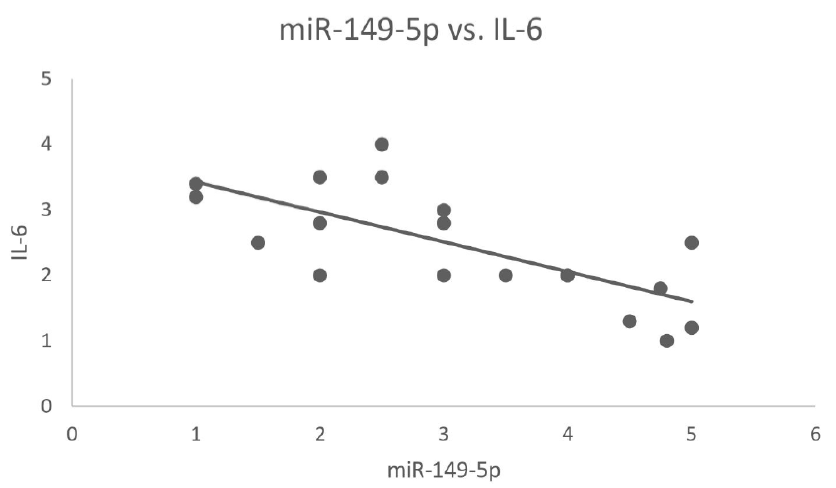
qRT-PCR gene expression analysis revealed the levels of miR-149-5p and IL-6 in blood samples from CKD patients, which were compared with those of normal individuals. When compared, CKD patients, particularly individuals with hypothyroidism, showed significant down-regulation in miR-149-5p levels and an increase in IL-6 levels. These findings suggest a potential role for miR-149-5p as well as IL-6 in CKD, which is to be further investigated. Figure 2 and Figure 3 depict the differential gene expression of miR-149-5p and IL-6 in CKD and normal blood samples. The relationship between increased IL-6 levels and decreased miR-149-5p levels in CKD patients suggests an inverse or negative correlation (as shown in Figure 4), thereby suggesting a systematic relationship between the two, with changes in one being associated with opposite changes in the other.
Discussion
CKD is a major threat to public health, impacting millions of individuals, as it represents a progressive and irreversible decline in kidney function that can ultimately progress to end-stage renal disease (ESRD). Inflammation, marked by an increase in pro-inflammatory cytokine levels such as IL-6, has a key role in the genesis of CKD. CKD is characterized by a chronic inflammatory state and increased oxidative stress, both of which contribute to disease progression and complications. Simultaneously, the dysregulation of key signaling pathways, such as NFκB, Wnt/β-catenin, and TGF-β, is implicated in CKD pathophysiology. These pathways are associated with inflammation, fibrosis, and abnormal cellular responses in the kidneys. Studies indicate that miR-149-5p frequently targets these signaling pathways8, 9, 10. Moreover, miR-149-5p has the potential to impact the cellular response to oxidative stress and inflammatory processes within the kidneys. By modulating these pathways, miR-149-5p may influence the intricate balance between inflammation and oxidative stress, contributing to the overall progression of CKD. Moreover, the observed relationship between elevated IL-6 levels and decreased miR-149-5p levels in individuals with CKD implies the presence of an inverse correlation. Such a correlation holds significance as it highlights a reciprocal influence between IL-6 and miR-149-5p levels, shedding light on potential regulatory mechanisms or interactions within the context of CKD.
Understanding these relationships provides valuable insights into the in tricate molecular dynamics associated with CKD pathology and may have implications for diagnostic or therapeutic approaches targeting these molecular components. Understanding theroleofmiR-149-5p in inflammation and oxidative stress holds significant clinical relevance in the context of CKD. Moreover, insights into miR-149-5p’s regulatory role could in form the development of targeted therapeutic interventions, aiming to modulate inflammation and oxidative stress to slow CKD progression. The clinical implications extend to personalized medicine, where understanding miR-149-5p’s involvement may lead to tailored interventions based on the specific molecular mechanisms underlying inflammation and oxidative stress in individual CKD patients. Overall, deciphering theroleofmiR-149-5p in CKD pathophysiology offers a promising avenue for advancing diagnostic and therapeutic strategies, ultimately improving patient outcomes in this prevalent and challenging condition.
Conclusions
In this study, we comprehensively assessed miR-149-5p and IL-6 expression in CKD patients compared to a control group of healthy individuals. Gaining a profound understanding of these molecular components holds the promise of pioneering new pharmaceutical strategies and enhancing therapeutic options for this prevalent condition, ultimately improving patient care for those with renal issues and consequently reducing morbidity and mortality rates.
Abbreviations
CKD: Chronic kidney disease; miRNA: microRNA; IL-6: Interleukin 6; ncRNA: non-coding RNA; NCBI: National Centre for Biotechnology Information; BLAST: Basic Local Alignment Search Tool; dNTP: deoxyribonucleotide triphosphates; ESRD: end-stage renal disease
Acknowledgments
Sekar D is a recipient of the Extramural Grants (2019-0106/CMB/ ADHOC/BMS and 5/4/8- 18/CD/2021-NCD-II), Indian Council of Medical Research (ICMR), Government of India, and their support is duly acknowledged.
Author’s contributions
Formal analysis, D.S; writing - original draft preparation, M.A; writing – review and editing; A.P; editing – revising; A.P. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Amaral
S.,
Hwang
W.,
Fivush
B.,
Neu
A.,
Frankenfield
D.,
Furth
S.,
Serum albumin level and risk for mortality and hospitalization in adolescents on hemodialysis. Clinical Journal of the American Society of Nephrology ; CJASN.
2008;
3
(3)
:
759-67
.
View Article PubMed Google Scholar -
Naugler
W.E.,
Sakurai
T.,
Kim
S.,
Gender Disparity in Liver Cancer Due to Sex Differences in MyD88-Dependent IL-6 Production. Science.
2007;
317
(5834)
:
121-124
.
View Article Google Scholar -
Nguyen
D.,
Ping
F.,
Mu
W.,
Hill
P.,
Atkins
R.C.,
Chadban
S.J.,
Macrophage accumulation in human progressive diabetic nephropathy. Nephrology (Carlton, Vic.).
2006;
11
(3)
:
226-31
.
View Article PubMed Google Scholar -
Yamashiro
H.,
Siomi
M.C.,
PIWI-Interacting RNA in Drosophila: Biogenesis, Transposon Regulation, and Beyond. Chemical Reviews.
2018;
118
(8)
:
4404-21
.
View Article PubMed Google Scholar -
Chen
C.,
Li
Y.,
Lu
H.,
Liu
K.,
Jiang
W.,
Zhang
Z.,
Curcumin attenuates vascular calcification via the exosomal miR-92b-3p/KLF4 axis. Experimental Biology and Medicine (Maywood, N.J.).
2022;
247
(16)
:
1420-32
.
View Article PubMed Google Scholar -
Li
H.,
Jia
Z.,
Li
A.,
Jenkins
G.,
Yang
X.,
Hu
J.,
Resveratrol repressed viability of U251 cells by miR-21 inhibiting of NF-κB pathway. Molecular and Cellular Biochemistry.
2013;
382
(1-2)
:
137-43
.
View Article PubMed Google Scholar -
Huang
X.,
Wang
L.,
Liu
W.,
Li
F.,
MicroRNA-497-5p inhibits proliferation and invasion of non-small cell lung cancer by regulating FGF2. Oncology Letters.
2019;
17
(3)
:
3425-31
.
View Article PubMed Google Scholar -
Ren
F.,
Yao
Y.,
Cai
X.,
Cai
Y.,
Su
Q.,
Fang
G.,
Ren F jia, Yao Y, Cai X yu, Cai Y ting, Su Q, Fang G ying. MiR-149-5p: An Important miRNA Regulated by Competing Endogenous RNAs in Diverse Human Cancers. Frontiers in Oncology.
2021;
11
:
743077
.
View Article Google Scholar -
Hübner
K.,
Karwelat
D.,
Pietsch
E.,
Beinborn
I.,
Winterberg
S.,
Bedenbender
K.,
NF-κB-mediated inhibition of microRNA-149-5p regulates Chitinase-3-like 1 expression in human airway epithelial cells. Cellular Signalling.
2020;
67
:
109498
.
View Article PubMed Google Scholar -
Ren
F.J.,
Yao
Y.,
Cai
X.Y.,
Cai
Y.T.,
Su
Q.,
Fang
G.Y.,
MiR-149-5p: An Important miRNA Regulated by Competing Endogenous RNAs in Diverse Human Cancers. Frontiers in Oncology.
2021;
11
:
743077
.
View Article PubMed Google Scholar -
Kreiner
F.F.,
Kraaijenhof
J.M.,
von Herrath
M.,
Hovingh
G.K.,
von Scholten
B.J.,
Interleukin 6 in diabetes, chronic kidney disease, and cardiovascular disease: mechanisms and therapeutic perspectives. Expert Review of Clinical Immunology.
2022;
18
(4)
:
377-89
.
View Article PubMed Google Scholar -
Saravanan
S.,
Islam
V.I.,
Thirugnanasambantham
K.,
Sekar
D.,
In Silico Identification of Human miR 3654 and its Targets Revealed its Involvement in Prostate Cancer Progression. MicroRNA (Shariqah, United Arab Emirates).
2016;
5
(2)
:
140-5
.
View Article PubMed Google Scholar -
Parvathy Sankar, Parvathy Sankar, Joscilin Mathew. Physiology, Blood Plasma.; 2023.
.
-
Krishnan
R.,
Mani
P.,
Sivakumar
P.,
Gopinath
V.,
Sekar
D.,
Expression and methylation of circulating microRNA-510 in essential hypertension. Hypertension Research.
2017;
40
(4)
:
361-3
.
View Article PubMed Google Scholar -
Rajkumar
K.V.,
Lakshmanan
G.,
Sekar
D.,
Identification of miR-802-5p and its involvement in type 2 diabetes mellitus. World Journal of Diabetes.
2020;
11
(12)
:
567-71
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 10 No 12 (2023)
Page No.: 6103-6109
Published on: 2023-12-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 3405 times
- PDF downloaded - 1283 times
- XML downloaded - 124 times
 Biomedpress
Biomedpress





