Abstract
Background: Polycystic ovarian syndrome (PCOS) cases have recently increased drastically among women during ovulation. The etiology of this endocrine disorder remains complex due to its multiple links that affect women of all ethnicities and races. Recent studies have implicated tumor necrosis factor-alpha (TNF-α) in PCOS pathophysiology. This study examines the associations of TNF-α polymorphisms 238G/A, 308G/A, and 1031T/C with PCOS.
Methods: We searched the Google Scholar, PubMed, EMBASE, Scopus, and Science Citation Index databases to identify suitable case-control studies and literature reviews for the statistical analysis. The obtained data were evaluated using the Review Manager 5.4 software. An odds ratio and 95% confidence interval were calculated for each genetic model.
Results: Twenty-three studies met the eligibility criteria, comprising 3294 cases and 3288 controls. Meta-analysis showed no significant association between TNF-α polymorphisms 238G/A and 308G/A and PCOS risk. However, TNF-α polymorphism 1031T/C was significantly associated with PCOS risk.
Conclusion: This meta-analysis indicates that TNF-α polymorphisms 238G/A and 308G/A may not be associated with PCOS risk, while TNF-α polymorphism 1031T/C appears associated with PCOS risk. However, a larger sample size is required to evaluate this association.
Introduction
Tumor necrosis factor-alpha (TNF-α), frequently found bound to the promoter region of genes implicated in various diseases, is a cytokine that promotes inflammation. Its gene is located in the class III region of the histocompatibility complex at chromosome 6p21.3, encoding a 157 amino acid (17 kDa) protein that forms a homotrimer1. It helps activate various inflammatory molecules, such as chemokines and cytokines. TNF-α greatly contributes to cellular homeostasis, differentiation, proliferation, and immune responses and regulates metabolite function in the body. It also helps in various biological activities such as enhancing neutrophil phagocytic ability and preventing liver cells from producing acute phase proteins, inhibiting or destroying tumor cells and viral replication2. Therefore, the dysregulated production or function of TNF-α causes various inflammatory diseases, including inflammatory bowel diseases, systemic lupus, multiple sclerosis, and rheumatoid arthritis. TNF-α is also known to induce hemorrhagic necrosis in murine Meth A sarcomas3. Around 43 single nucleotide polymorphisms (SNPs) have been identified in the promotor region of TNF-α (https://shorturl.at/ioDX9). However, some studies have reported conflicting results regarding their association with changes in TNF-α levels.
TNF-α has soluble and transmembrane forms that bind to outer membrane-bound receptors on the target cells. Specifically, the membrane-bound metalloproteinase TNF-α converting enzyme (TACE) is required to synthesize soluble TNF-α from transmembrane TNF-α4. TNF-α binds to type-1 and type-2 receptors. Type-1 receptors are TNF receptor superfamily member 1A (TNFRSF1A/TNFR1/CD120a) and CD4 molecule (CD4/p55)5. Type-2 receptors are TNFRSF1A and TNF receptor superfamily member 1B (TNFRSF1B/TNFR2/p75/CD120b). The TNFR1 receptor is important in regulating inflammatory pathways and is mainly expressed in human tissues. The TNFR2 receptor greatly affects tumor cell development by promoting immune escape6. Both TNFR1 and TNFR2 receptors induce cellular signals for biological activities, including inflammation and cell death. Several distinct signaling complexes designated as I, IIa, IIb, and IIc all induce unique cell responses. These responses are controlled by the transmembrane and soluble forms of TNF-α, which bind to the death domain adaptor protein to trigger cell apoptosis or growth via the TNFR1 receptor7.
During complex I assembly, TNFR1 stimulates and attaches to the TNFR1-associated death domain (TRADD) protein, followed by the assembly and cooperation of many parts, including TNF receptor-associated factors 2 (TRAF2) and 5 (TRAF5), ubiquitin-conjugating enzyme E1 (UBE1), receptor-interacting threonine/serine protein kinase 1 (RIPK1), and cellular inhibitor of apoptosis proteins 1 (cIAP1) and 2 (cIAP2). Therefore, complex I activates the nuclear factor kappa B (NF-κB) and mitogen-activated protein kinase (MAPK) pathways, leading to cell proliferation, tissue degeneration, cell survival, and inflammation8.
Unlike complex I, complexes IIa, IIb, and IIc are assembled in the cytoplasm rather than at the plasma membrane. Pro-Caspase 8 (pro-CASP8), Fas-associated protein with death domain (FADD), TRAF2, RIPK1, and cIAP1/2 are all components of complex IIa. The complex IIb components are arranged in the same manner as those of complex IIa, except for a protein called receptor-interacting serine/threonine kinase 3 (RIPK3)9. An adaptor protein complex, called the apoptosome, helps complexes IIa and IIb induce apoptosis by aiding CASP8 activation. Necrosomal complex IIc is generated when RIPK1 and RIPK3 bind without being cleaved. This complex triggers necroptosis and inflammation by activating mixed lineage kinase domain (MLKL)10. In general, cell stimulation, migration, and replication are predominantly triggered by TNFR2, whereas cytotoxicity and inflammation are triggered by TNFR1.
TNF-α is known for its crucial role in autoimmune disorders, including psoriasis, noninfectious uveitis (NIU), psoriatic arthritis, and rheumatoid arthritis. Psoriatic arthritis affects 1% of the population, with physical characteristics such as swollen toes and fingers and inflamed joints. Activated dendritic cells (DCs), T helper 17 (Th17) cells, and macrophages are primarily involved in the pathogenesis of psoriatic arthritis, which is triggered by the overproduction of interleukin (IL)-23 and TNF-α11, 12. IL-23 promotes the differentiation of naive T cells into Th17 cells, which overproduce IL-17. Then, inflammatory cells such as DCs become activated after stimulation by TNF-α and IL-17. TNF-α promotes anti-apoptosis and keratinocyte proliferation through the transforming growth factor (TNF)-ß signaling pathway, increasing the recruitment of inflammatory cells and resulting in the formation of microabscesses in psoriasis13. The psoriasis lesions are mainly subdivided into five types: guttate, erythrodermic, inverse, plaque, and pustular. The dysregulation of skin immune responses is reflected by angiogenesis and epidermal hyperplasia on the lesions14. Most clinical psoriasis subtypes have a common inflammatory mechanism that contributes to psoriasis development15. DCs become activated by IL-12 and induce differentiation of IL-23 into Th17 cells and naive T cells into T helper 1 (Th1) cells, which secretes TNF-α and interferon (IFN), while Th17 cells secrete abundant IL-17. Therefore, epidermal alterations and keratinocyte hyperproliferation, including hypogranulosis, parakeratosis, and acanthosis, are caused by TNF-α, IFN-γ, and IL-1716.
Another autoimmune disorder related to the eye is NIU17. In addition to causing blindness or visual impairment, NIU has been associated with the development of visual distribution, cataracts, retinal detachment, and glaucoma. Many cytokines, including IL-10, IL-12, IL-23, and IL-6, are generated by macrophages18. TNF-α and other cytokines help activate DCs. Excess IL-12 production by activated DCs causes naive T cells to differentiate into Th1 cells19. Overproduction of IL-6 and TGF-β by DCs is a key factor in Th17 cell development. Activated Th1 and Th17 cells penetrate the choroid layers that supply blood to the retina. Migration of Th1 and Th17 cells can stimulate the retinal vasculature, attracting nonspecific blood-circulating leukocytes. NIU reflects inflammation that causes uvea destruction and leads to edema20.
In the modern lifestyle, many women of premenopausal age are affected by polycystic ovary syndrome (PCOS) caused by endocrine disorders associated with genetic factors such as premature fetal development, early follicle maturity, and a family history of PCOS. It occurs in about 1 in 13 premenopausal women21. Stein and Leventhal first described PCOS in 1935 in a female patient with oligo-ovulatory infertility22. This condition is mainly caused by altered ovulation and hyperandrogenism, leading to complications such as ovarian enlargement, infertility, endometrial cancer, and other diseases. In India, its prevalence is about 8.25%–22.5% based on lifestyle and food habits23. In 1990, diagnostic criteria were produced for PCOS at conferences funded by the National Institutes of Child and Health (NICH) and Human Development (HD)24. In 2003, Rotterdam proposed classification criteria at the conference organized by the American Society of Reproductive Medicine (ASRM) and the European Society of Human Reproduction and Embryology (ESHRE), which was primarily used to classify PCOS25. Another conference organized by the PCOS Society in 2006 proposed diagnostic features for PCOS26. Individuals affected by PCOS are mostly obese due to androgen overexpression, which increases adipose tissue in the abdominal region. The classification criteria proposed at various conferences are listed in Table 1.
| Year Proposed | Proposed By | Features |
|---|---|---|
| 1990 | NICH and HD | Hyperandrogenism, oligo-ovulation, thyroid, hyperprolactinemia, and inherited adrenal hyperplasia |
| 2003 | ASRM and ESHRE | Hyperandrogenism and oligo-ovulation |
| 2006 | Androgen and PCOS Societies | Clinical and biochemical analysis of hyperandrogenism and oligo-ovulation |
Genic SNPs and single nucleotide variants influence steroidogenesis, ovarian theca cell activity, and the release of hormones from the hypothalamus and pituitary gland27, 28. Epigenetic factors such as intrauterine exposure and excess androgen in the maternal environment can also cause stable, heritable phenotypes that contribute to PCOS. Hyperandrogenism, abnormal steroid production, insulin resistance, and central obesity are all symptoms of a malfunctioning hypothalamic-pituitary-ovarian axis, which results in PCOS. Hyperandrogenism is caused by excess androgen secretion by the theca cells in the ovaries in response to adipose tissue development, leading to the formation of small multiple antral follicles and a sex hormone imbalance, causing endometrial carcinoma. Oocyte quality and endothelial function are both adversely affected by chronic oxidative stress and proinflammatory cytokines, which indicate infertility. However, prescreening and diagnosis are crucial in preventing PCOS and helping prevent metabolic abnormalities. Physical and mental well-being and a healthy lifestyle and environment play significant roles in overcoming the PCOS burden.
Several studies have looked at the association of TNF-α with various autoimmune diseases. However, SNP-based studies have been limited to only one or two genes, and their results have been limited29. Qualified data still needs to be incorporated to improve results. It has been demonstrated that TNF-α levels are elevated in the serum and follicular fluid of women with PCOS. Therefore, this study aimed to assess the associations of TNF-α polymorphisms 1031T/C, 308G/A, and 238G/A with PCOS.
Methods
Literature search
A literature search was conducted in all available public and scientific databases, including Embase, NCBI, Google Scholar, Medline, and Science Direct, from inception to April 2023 using the following keywords to identify all articles on the association of 238G/A, 308G/A, and 1031 T/C with PCOS: TNF-alpha, TNF-alpha with polycystic ovarian syndrome, PCOS-TNF-alpha, and PCOS. Only English-language articles were considered. In order to avoid duplicated studies, authors’ names were searched and screened in all the databases to identify appropriate studies, titles, abstracts, and full texts. In addition, the reference lists of the identified articles were also screened.
Selection criteria
The studies had to meet all of the following inclusion criteria: (i) evaluate the relationship between the TNF-α 238G/A, 308G/A, and 1031T/C polymorphisms and PCOS; (ii) include patients with PCOS and controls with eligible genotypic and phenotypic distributions of TNF-α 238G/A, 308G/A, and 1031T/C polymorphisms; (iii) both cases and controls are of the same ethnicity; and (iv) the full text was available in English. The exclusion criteria were as follows: (i) no control group; (ii) low 95% confidence interval or odds ratio (OR); (iii) studies with overlapping data; and (iv) animal studies. Most studies on Caucasians have not examined the association between PCOS and TNF-α 238G/A, 308G/A, and 1031T/C polymorphisms. We identified very few studies on Caucasians, and many were excluded due to the lack of proper inclusion criteria.
Data extraction
The following data were extracted from the articles selected for inclusion: publication year, first author, origin, ethnicity, number of cases, genotyping methods, and controls registered. The conflict and disagreement from the selected articles were removed from the study.
Statistical analysis
The data were analyzed using the Review Manager 5.4 and MetaGenyo software. In order to determine whether the study was significant, we determined whether the p-value was significant at p < 0.005 using genetic variations such as allele comparison, dominant regression, over-dominant, and recessive. The consistency of the findings across all studies was evaluated using the inconsistency index (I2), which ranges from 0 to 100. The inconsistency index is crucial in determining the homogeneity (0% significance) and heterogeneity indications, which are responsible for most variations30. The degree of heterogeneity among the studies was assessed using Q-statistics and the chi-square test. The z-test was used to calculate ORs, and shared results among studies were considered statistically significant at p < 0.05. A sensitivity analysis was performed to assess the relative contributions of the included studies to the total estimates, removing one study at a time. Funnel plots and Egger’s linear regression test were used to detect publication bias. The log standard error was plotted against the odds ratios for each study. The heterogeneity between the eligible analyses performed using Egger’s test, Q-test, and inconsistency index statistics was considered statistically significant if p < 0.005.
| Contents | Study | Ethnicity | AA_Cases/Controls | GA_Cases/Controls | GG_Cases/Controls | Total Cases/Controls | Hardy-Weinberg equilibrium p-value |
|---|---|---|---|---|---|---|---|
| TNF-Alpha 238G/A | Sampurna et al . 2021 31 | Asian | 32/30 | 44/45 | 24/25 | 100/100 | 0.3283 |
| Bhatnagar et al . 2019 32 | Asian | 2/0 | 50/83 | 148/117 | 200/200 | 0.0002 | |
| Kordestani et al . 2018 33 | Asian | 1/3 | 6/6 | 104/96 | 111/105 | 0 | |
| Wen et al . 2013 34 | Asian | 0/0 | 7/4 | 137/68 | 144/72 | 0.8084 | |
| Xie et al . 2016 35 | Asian | 0/0 | 3/3 | 99/93 | 102/96 | 0.8764 | |
| Kordestani et al. 2018 33 | Asian | 1/3 | 6/6 | 104/96 | 111/105 | 0 | |
| TNF-Alpha 308G/A | Sampurna et al . 2021 31 | Asian | 32/30 | 44/45 | 24/25 | 100/100 | 0.3283 |
| Bhatnagar et al . 2019 32 | Asian | 2/0 | 50/83 | 148/117 | 200/200 | 0.0002 | |
| Sampurna et al . 2021 31 | Asian | 25/30 | 50/47 | 25/23 | 100/100 | 0.5798 | |
| Azeez et al . 2021 36 | Asian | 1/1 | 25/15 | 32/14 | 58/30 | 0.2054 | |
| Alwan et al. 2021 37 | Asian | 9/7 | 15/13 | 56/50 | 80/70 | 0.0007 | |
| Li et al. 2017 38 | Asian | 3/1 | 33/24 | 357/356 | 393/381 | 0.387 | |
| Bhatnagar et al . 2019 32 | Asian | 0/2 | 79/63 | 121/135 | 200/200 | 0.0671 | |
| Deepika et al . 2013 39 | Asian | 3/3 | 10/10 | 270/293 | 283/306 | 0 | |
| Mao et al . 2000 40 | Asian | 1/4 | 29/13 | 88/37 | 118/54 | 0.0889 | |
| Milner et al. 1999 41 | Caucasian | 2/3 | 23/42 | 59/63 | 84/108 | 0.1939 | |
| Peng et al . 2010 42 | Asian | 1/2 | 11/27 | 118/146 | 130/175 | 0.557 | |
| Vural et al . 2010 43 | Asian | 3/3 | 16/15 | 78/77 | 97/95 | 0.0549 | |
| Wen et al . 2013 34 | Asian | 0/0 | 14/7 | 89/52 | 103/59 | 0.6281 | |
| TNF-Alpha 1031T/C | Sampurna et al. 2021 31 | Asian | 38/35 | 38/40 | 24/25 | 100/100 | 0.055 |
| Bhatnager et al. 2019 32 | Asian | 0/2 | 48/69 | 152/129 | 200/200 | 0.0272 | |
| Deepika et al. 2013 39 | Asian | 6/5 | 170/139 | 107/162 | 283/306 | 0 | |
| Yun et al . 2011 44 | Asian | 2/0 | 71/22 | 144/122 | 217/144 | 0.321 | |
| Hazwanie et al . 2015 45 | Asian | 3/91 | 8/45 | 1/9 | 12/145 | 0.2922 | |
| Alkhuriji et al. 2020 46 | Asian | 1/9 | 17/46 | 50/82 | 68/137 | 0.4666 |
Results
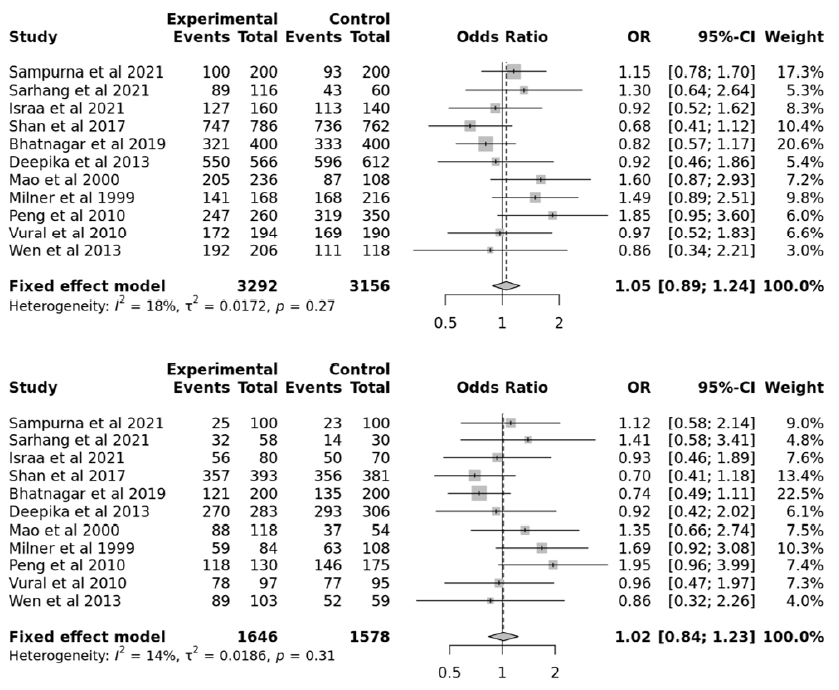
This study aimed to identify associations between TNF-α gene polymorphisms and PCOS. The searches of the various databases, including Google Scholar, NCBI, and Science Direct, identified six studies with 768 cases and 678 controls for the TNF-α 238G/A polymorphism47, 32, 33, 31, 34, 35, 11 studies with 1646 cases and 1578 controls for the TNF-α 308G/A polymorphism32, 31, 34, 39, 48, 41, 36, 38, 43, 37, 40, and six studies with 880 cases and 1032 controls for the TNF-α 1031T/C polymorphism32, 31, 39, 46, 45, 44. Based on the data collected from these 23 studies, PCOS was associated with the TNF-α 238G/A, 308G/A, and 1031T/C polymorphism (Figure 1). The data selected for the analysis is shown in Table 2. Among the selected studies, 22 were conducted in Asians and one in Caucasians. Begg’s funnel plot, funnel plot, and Egger’s test were performed for statistical evidence, and heterogeneity was observed from all selected articles.
| Model | Ethnicity | Number of studies | Test of association | Test of heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|---|
| - | Odd ratio | 95% confidence interval | p-value | Model | p-value | I 2 | p-value (Egger's test) | ||
| Allele contrast (A vs . a) | Overall | 6 | 1.2860 | [1.0001; 1.6537] | 0.049953 | Fixed | 0.3215 | 0.1457 | 0.957 |
| Recessive model (AA vs . Aa+aa) | Overall | 6 | 1.5287 | [1.1134; 2.0989] | 0.008691 | Fixed | 0.3803 | 0.0463 | 0.2936 |
| Dominant model (AA+Aa vs . aa) | Overall | 3 | 0.9344 | [0.5287; 1.6516] | 0.815448 | Fixed | 0.3419 | 0.0683 | 0.8213 |
| Overdominant (Aa vs. AA + aa) | Overall | 5 | 0.6539 | [0.4800; 0.8908] | 0.007073 | Fixed | 0.2978 | 0.1835 | 0.3097 |
| Model | Ethnicity | Number of studies | Test of association | Test of heterogeneity | Publication bias | ||||
|---|---|---|---|---|---|---|---|---|---|
| - | - | Odd ratio | 95% confidence interval | p-value | Model | p-value | I 2 | p-value (Egger's test) | |
| Allele contrast (A vs . a) | Overall | 11 | 1.0494 | [0.8912; 1.2356] | 0.562921 | Fixed | 0.2733 | 0.1789 | 0.3883 |
| Recessive model (AA vs . Aa+aa) | Overall | 11 | 1.0159 | [0.8364; 1.2338] | 0.873805 | Fixed | 0.3078 | 0.143 | 0.1283 |
| Dominant model (AA+Aa vs . aa) | Overall | 10 | 1.2340 | [0.8015; 1.9000] | 0.339546 | Fixed | 0.7397 | 0 | 0.4684 |
| Overdominant (Aa vs . AA + aa) | Overall | 11 | 1.0380 | [0.8515; 1.2654] | 0.711939 | Fixed | 0.3892 | 0.057 | 0.1445 |
| Model | Ethnicity | Number of studies | Test of association | Test of heterogeneity | Publication bias | ||||
|---|---|---|---|---|---|---|---|---|---|
| - | - | Odd ratio | 95% confidence interval | p-value | Model | p-value | I 2 | p-value (Egger's test) | |
| Allele contrast (A vs . a) | Overall | 6 | 1.0641 | [0.6570; 1.7235] | 0.800633 | Random | 0 | 0.8621 | 0.3096 |
| Recessive model (AA vs. Aa+aa) | Overall | 6 | 0.912 | [0.4971; 1.6733] | 0.766151 | Random | 0 | 0.8524 | 0.5966 |
| Dominant model (AA+Aa vs. aa) | Overall | 6 | 1.5185 | [0.6634; 3.4755] | 0.322802 | Random | 0.097 | 0.4635 | 0.3609 |
| Overdominant (Aa vs . AA + aa) | Overall | 6 | 1.3060 | [0.7446; 2.2907] | 0.35179 | Random | 0 | 0.8456 | 0.9502 |
Quantitative data analysis
The meta-analysis included 23 total studies to assess the association of PCOS with TNF-α 238G/A, 308G/A, and 1031T/C polymorphisms. We combined all the collected data with 3288 controls and 3294 cases. The TNF-α 238G/A polymorphism showed no significant associations in the allelic, recessive, dominant, and over-dominant models (p > 0.05; Table 3). In addition, subgroup analyses with allelic, recessive, dominant, and over-dominant models were nonsignificant (p > 0.05). Similarly, the TNF-α 308G/A polymorphism showed no significant associations in the allelic, dominant, overdominant, and recessive models (p > 0.05; Table 4). Again, subgroup analyses with the allelic, dominant, recessive, and over-dominant models were nonsignificant (p > 0.05). However, the TNF-α 1031T/C polymorphism showed significant associations with the allelic, recessive, and over-dominant models (p < 0.05; Table 5).
Publication Bias
Each variable was checked for apparent publication bias due to sample size constraints and reporting bias. A forest plot represented the heterogenicity, and a sensitivity analysis was conducted to identify studies contributing to the total estimates. Statistically constant results were obtained from the collected data (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7), and significant results (Figure 8, Figure 9, Figure 10) were obtained when the funnel plot was statistically analyzed (Figure 11, Figure 12, Figure 13).
Discussion
PCOS is the most commonly occurring endocrine disorder among adult women. In PCOS, ovarian dysfunction causes other metabolic disorders, which may result in chronic inflammation. TNF-α, an inflammatory cytokine found in the human ovaries, ovarian follicular fluid, and oocytes, causes many inflammatory disorders and impacts follicular atresia, physiological ovulation dysfunction, ovarian apoptosis, anovulation, and steroid secretion. The pathophysiological and etiological mechanisms of PCOS are highly complex. Inflammation and immune-regulatory genes are responsible for PCOS development and progression. However, many studies have not succeeded in identifying a significant relationship between inflammatory pathways and PCOS development.
Our study comprised 2764 cases with PCOS and 3218 controls obtained from published studies exploring associations between PCOS and TNF-α 238G/A, 308G/A, and 1031T/C polymorphisms. In our study, the TNF-α 238G/A polymorphism showed no association with PCOS in all genetic models (allele, recessive, over-dominant, and dominant), suggesting that TNF-α 238G/A may not contribute to PCOS development. There is no conclusive proof associating the TNF-α 308G/A polymorphism with PCOS. However, the TNF-α 1031T/C polymorphism was significantly associated with PCOS in the allelic, recessive, and over-dominant models but not in the dominant model. Therefore, these results suggest that the TNF-α 1031T/C polymorphism may not be associated with PCOS.
Our meta-analysis has generated some important findings, but it is not without limitations. Since we only considered research studies published in English that were abstracted and indexed in online databases, we might have missed relevant studies published in other languages. We also did not investigate the possible influence of gene-environment interactions, and a subgroup analysis was not performed because there was a dearth of appropriate studies and data. Nonetheless, our meta-analysis does have some strengths. Firstly, our qualitative and quantitative analysis, such as funnel plot and Egger’s linear regression, did not demonstrate clear evidence of publication bias. Therefore, we can conclude that our findings have strong statistical support. Secondly, a strict data extraction and analysis procedure was performed to draw satisfactory and reliable conclusions from the study.
Conclusions
In conclusion, our meta-analysis assessed possible associations of TNF-α 238G/A, 308G/A, and 1031T/C polymorphisms with PCOS using valuable statistical data from significant and nonsignificant studies. Overall, our meta-analysis showed that the TNF-α 238G/A and 308G/A polymorphisms may not be associated with PCOS, whereas the TNF-α 1031T/C polymorphism may be associated with PCOS. Future studies should explore possible associations and interactions between TNF-α polymorphisms and PCOS using large datasets comprising gene and environment data to confirm our findings.
Abbreviations
PCOS: Polycystic ovarian syndrome, TNF-α: Tumor necrosis factor-alpha, SNPs: single nucleotide polymorphisms, TACE: TNF-α converting enzyme
Acknowledgments
Gowtham, Harini, Thirumalai, and Pragya acknowledge CARE for fellowship.
Author’s contributions
All authors significantly contributed to this work, read and approved the final manuscript.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Jang
D.I.,
Lee
A.H.,
Shin
H.Y.,
Song
H.R.,
Park
J.H.,
Kang
T.B.,
The role of tumor necrosis factor alpha (TNF-α) in autoimmune disease and current TNF-α inhibitors in therapeutics. International Journal of Molecular Sciences.
2021;
22
(5)
:
2719
.
View Article PubMed Google Scholar -
Hong
L.,
Zhang
Y.,
Wang
Q.,
Han
Y.,
Teng
X.,
Effects of interleukin 6 and tumor necrosis factor-α on the proliferation of porcine theca interna cells: possible role of these cytokines in the pathogenesis of polycystic ovary syndrome. Taiwanese Journal of Obstetrics & Gynecology.
2016;
55
(2)
:
183-7
.
View Article PubMed Google Scholar -
Li
X.,
Körner
H.,
Liu
X.,
Susceptibility to intracellular infections: contributions of TNF to immune defense. Frontiers in Microbiology.
2020;
11
:
1643
.
View Article PubMed Google Scholar -
Jiang
Y.,
Yu
M.,
Hu
X.,
Han
L.,
Yang
K.,
Ba
H.,
STAT1 mediates transmembrane TNF-alpha-induced formation of death-inducing signaling complex and apoptotic signaling via TNFR1. Cell Death and Differentiation.
2017;
24
(4)
:
660-71
.
View Article PubMed Google Scholar -
Assrawi
E.,
Louvrier
C.,
El Khouri
E.,
Delaleu
J.,
Copin
B.,
Dastot-Le Moal
F.,
Mosaic variants in TNFRSF1A: an emerging cause of tumour necrosis factor receptor-associated periodic syndrome. Rheumatology (Oxford, England).
2022;
62
(1)
:
473-9
.
View Article PubMed Google Scholar -
Yang
Y.,
Islam
M.S.,
Hu
Y.,
Chen
X.,
TNFR2: role in cancer immunology and immunotherapy. ImmunoTargets and Therapy.
2021;
10
:
103-22
.
View Article PubMed Google Scholar -
Pobezinskaya
Y.L.,
Liu
Z.,
The role of TRADD in death receptor signaling. Cell Cycle (Georgetown, Tex.).
2012;
11
(5)
:
871-6
.
View Article PubMed Google Scholar -
Shi
J.H.,
Sun
S.C.,
Tumor necrosis factor receptor-associated factor regulation of nuclear factor κB and mitogen-activated protein kinase pathways. Frontiers in Immunology.
2018;
9
:
1849
.
View Article PubMed Google Scholar -
Anderton
H.,
Bandala-Sanchez
E.,
Simpson
D.S.,
Rickard
J.A.,
Ng
A.P.,
Di Rago
L.,
RIPK1 prevents TRADD-driven, but TNFR1 independent, apoptosis during development. Cell Death and Differentiation.
2019;
26
(5)
:
877-89
.
View Article PubMed Google Scholar -
Holbrook
J.,
Lara-Reyna
S.,
Jarosz-Griffiths
H.,
McDermott
M.,
Tumour necrosis factor signalling in health and disease. F1000 Research.
2019;
8
(111)
:
111
.
View Article PubMed Google Scholar -
Veale
D.J.,
Fearon
U.,
The pathogenesis of psoriatic arthritis. Lancet.
2018;
391
(10136)
:
2273-84
.
View Article PubMed Google Scholar -
Affandi
A.J.,
Silva-Cardoso
S.C.,
Garcia
S.,
Leijten
E.F.,
van Kempen
T.S.,
Marut
W.,
CXCL4 is a novel inducer of human Th17 cells and correlates with IL-17 and IL-22 in psoriatic arthritis. European Journal of Immunology.
2018;
48
(3)
:
522-31
.
View Article PubMed Google Scholar -
Ogawa
E.,
Sato
Y.,
Minagawa
A.,
Okuyama
R.,
Pathogenesis of psoriasis and development of treatment. The Journal of Dermatology.
2018;
45
(3)
:
264-72
.
View Article PubMed Google Scholar -
Liu
L.,
Sun
X.Y.,
Lu
Y.,
Song
J.K.,
Xing
M.,
Chen
X.,
Fire needle therapy for the treatment of psoriasis: a quantitative evidence synthesis. Journal of Alternative and Complementary Medicine (New York, N.Y.).
2021;
27
(1)
:
24-37
.
View Article PubMed Google Scholar -
Katsimbri
P.,
Korakas
E.,
Kountouri
A.,
Ikonomidis
I.,
Tsougos
E.,
Vlachos
D.,
The effect of antioxidant and anti-inflammatory capacity of diet on psoriasis and psoriatic arthritis phenotype: nutrition as therapeutic tool?. Antioxidants.
2021;
10
(2)
:
157
.
View Article PubMed Google Scholar -
Griffiths
C.E.,
Barker
J.N.,
Pathogenesis and clinical features of psoriasis. Lancet.
2007;
370
(9583)
:
263-71
.
View Article PubMed Google Scholar -
Rosenbaum JT, Bodaghi B, Couto C, Zierhut M, Acharya N, Pavesio C, et al., editors. New observations and emerging ideas in diagnosis and management of non-infectious uveitis: a review. Seminars in arthritis and rheumatism; 2019: Elsevier. .
View Article Google Scholar -
Ghasemi
H.,
Ghazanfari
T.,
Yaraee
R.,
Owlia
P.,
Hassan
Z.M.,
Faghihzadeh
S.,
Roles of IL-10 in ocular inflammations: a review. Ocular Immunology and Inflammation.
2012;
20
(6)
:
406-18
.
View Article PubMed Google Scholar -
Wu
X.,
Tian
J.,
Wang
S.,
Insight into non-pathogenic Th17 cells in autoimmune diseases. Frontiers in Immunology.
2018;
9
:
1112
.
View Article PubMed Google Scholar -
Fang
L.,
Liu
J.,
Liu
Z.,
Zhou
H.,
Immune modulating nanoparticles for the treatment of ocular diseases. Journal of Nanobiotechnology.
2022;
20
(1)
:
496
.
View Article PubMed Google Scholar -
Livadas
S.,
Anagnostis
P.,
Bosdou
J.K.,
Bantouna
D.,
Paparodis
R.,
Polycystic ovary syndrome and type 2 diabetes mellitus: A state-of-the-art review. World Journal of Diabetes.
2022;
13
(1)
:
5-26
.
View Article PubMed Google Scholar -
Hachey
L.M.,
Kroger-Jarvis
M.,
Pavlik-Maus
T.,
Leach
R.,
Clinical implications of polycystic ovary syndrome in adolescents. Nursing for Women{&}{#}x0027;s Health.
2020;
24
(2)
:
115-26
.
View Article PubMed Google Scholar -
Mehreen
T.S.,
Ranjani
H.,
Kamalesh
R.,
Ram
U.,
Anjana
R.M.,
Mohan
V.,
Prevalence of polycystic ovarian syndrome among adolescents and young women in India. Journal of Diabetology..
2021;
12
(3)
:
319-25
.
View Article Google Scholar -
Göbl
C.S.,
Ott
J.,
Bozkurt
L.,
Feichtinger
M.,
Rehmann
V.,
Cserjan
A.,
To assess the association between glucose metabolism and ectopic lipid content in different clinical classifications of PCOS. PLoS One.
2016;
11
(8)
:
e0160571
.
View Article PubMed Google Scholar -
Azziz
R.,
Controversy in clinical endocrinology: diagnosis of polycystic ovarian syndrome: the Rotterdam criteria are premature. The Journal of Clinical Endocrinology and Metabolism.
2006;
91
(3)
:
781-5
.
View Article PubMed Google Scholar -
Goodarzi
M.O.,
Dumesic
D.A.,
Chazenbalk
G.,
Azziz
R.,
Polycystic ovary syndrome: etiology, pathogenesis and diagnosis. Nature Reviews. Endocrinology.
2011;
7
(4)
:
219-31
.
View Article PubMed Google Scholar -
Veerabathiran
R.,
Srinivasan
K.,
Jayaprasad
P.,
Iyshwarya
B.,
Husain
R.A.,
Association of MTHFR gene polymorphism in preeclampsia and recurrent pregnancy loss: A case-control study from South India. Human Gene.
2023;
37
:
201199
.
View Article Google Scholar -
Varghese
S.,
Kumar
S.G.,
Role of eNOS and TGFβ1 gene polymorphisms in the development of diabetic nephropathy in type 2 diabetic patients in South Indian population. The Egyptian Journal of Medical Human Genetics.
2022;
23
:
1-10
.
-
Qi
X.,
Wang
X.Q.,
Jin
L.,
Gao
L.X.,
Guo
H.F.,
Uncovering potential single nucleotide polymorphisms, copy number variations and related signaling pathways in primary Sjogren's syndrome. Bioengineered.
2021;
12
(2)
:
9313-31
.
View Article PubMed Google Scholar -
Murugesan
L.,
Babu
K.,
Puthamohan
V.M.,
Basavaraju
P.,
Devaraj
I.,
Balasubramani
R.,
A review on genetic polymorphism in MTHFR gene with Down syndrome and leukemia. Meta Gene.
2020;
25
:
100752
.
View Article Google Scholar -
Alkhuriji
A.F.,
Omar
S.Y. Al,
Babay
Z.A.,
El-Khadragy
M.F.,
Mansour
L.A.,
Alharbi
W.G.,
Association of IL-1β, IL-6, TNF-α, and TGFβ1 gene polymorphisms with recurrent spontaneous abortion in polycystic ovary syndrome. Disease Markers.
2020;
2020
:
6076274
.
View Article Google Scholar -
Vural
P.,
Değirmencioğlu
S.,
Saral
N.Y.,
Akgül
C.,
Tumor necrosis factor α (-308), interleukin-6 (-174) and interleukin-10 (-1082) gene polymorphisms in polycystic ovary syndrome. European Journal of Obstetrics, Gynecology, and Reproductive Biology.
2010;
150
(1)
:
61-5
.
View Article PubMed Google Scholar -
Peng
C.Y.,
Long
X.Y.,
Lu
G.X.,
Association of AR rs6152G/A gene polymorphism with susceptibility to polycystic ovary syndrome in Chinese women. Reproduction, Fertility and Development.
2010;
22
(5)
:
881-885
.
View Article Google Scholar -
Milner
C.R.,
Craig
J.E.,
Hussey
N.D.,
Norman
R.J.,
No association between the -308 polymorphism in the tumour necrosis factor α (TNFalpha) promoter region and polycystic ovaries. Molecular Human Reproduction.
1999;
5
(1)
:
5-9
.
View Article PubMed Google Scholar -
Mao
W.,
Yu
L.,
Chen
Y.,
[Study on the relationship between a polymorphism of tumor necrosis factor-alpha gene and the pathogenesis of polycystic ovary syndrome]. Zhonghua Fu Chan Ke Za Zhi.
2000;
35
(9)
:
536-9
.
PubMed Google Scholar -
Hazwanie
H.,
Gan
S.,
Nalliah
S.,
The relevance of genetic polymorphism of CYP1A1, ICAM-1, TNF-α and INSR genes in women with polycystic ovary syndrome (PCOS). Journal of Medical and Bioengineering.
2015;
4
(5)
:
367-70
.
View Article Google Scholar -
Yun
J.H.,
Choi
J.W.,
Lee
K.J.,
Shin
J.S.,
Baek
K.H.,
The promoter -1031(T/C) polymorphism in tumor necrosis factor-alpha associated with polycystic ovary syndrome. Reproductive Biology and Endocrinology.
2011;
9
(1)
:
131
.
View Article PubMed Google Scholar -
Deepika
M.L.,
Reddy
K.R.,
Yashwanth
A.,
Rani
V.U.,
Latha
K.P.,
Jahan
P.,
TNF-α haplotype association with polycystic ovary syndrome - a South Indian study. Journal of Assisted Reproduction and Genetics.
2013;
30
(11)
:
1493-503
.
View Article PubMed Google Scholar -
Bhatnager
R.,
Jalthuria
J.,
Sehrawat
R.,
Nanda
S.,
Dang
A.S.,
Evaluating the association of TNF α promoter haplotype with its serum levels and the risk of PCOS: A case control study. Cytokine.
2019;
114
:
86-91
.
View Article PubMed Google Scholar -
Sampurna
K.,
Ramesh
B.,
Rajesh
B.,
Madhavi
S.,
Parveen
S.,
Bhargav
P.,
Genotype- Phenotype correlations of Leptin receptor gene in Polycystic ovarian disease (PCOS). IOSR Journal of Biotechnology and Biochemistry.
2018;
4
(1)
:
20-27
.
-
Wen
Q.,
Wu
J.,
Wu
L.,
Liu
L.,
Yang
H.,
Sun
Z.,
Association of TNF-α G-308A and G-238A gene polymorphisms with PCOS in women. Zhonghua Linchuang Yishi Zazhi.
2013;
7
:
3394-9
.
-
Kordestani
F.,
Mazloomi
S.,
Mortazavi
Y.,
Mazloomzadeh
S.,
Fathi
M.,
Rahmanpour
H.,
Preliminary study showing no association between G238A (rs361525) tumor necrosis factor-α (TNF-α) gene polymorphism and its serum level, hormonal and biochemical aspects of polycystic ovary syndrome. BMC Medical Genetics.
2018;
19
(1)
:
149
.
View Article PubMed Google Scholar -
Shi
X.,
Xie
X.,
Jia
Y.,
Li
S.,
Associations of insulin receptor and insulin receptor substrates genetic polymorphisms with polycystic ovary syndrome: A systematic review and meta-analysis. Journal of Obstetrics and Gynaecology Research.
2016;
42
(7)
:
844-54
.
View Article PubMed Google Scholar -
Azeez
S.H.,
Ismail
I.B.,
Darogha
S.N.,
The Effect of Interleukin-6 and Tumor Necrosis Factor-Alpha Gene Polymorphism and Hormone Replacement Therapy on Polycystic Ovary Syndrome. Cellular and Molecular Biology.
2022;
67
(5)
:
278-85
.
View Article PubMed Google Scholar -
Alwan
I.A.,
Al-Heety
R.A.,
Association of tumor necrosis factor-α (- 308 G/A) and interleukin-10 (- 1082 A/G) gene polymorphisms with polycystic ovary syndrome in Iraqi women. Meta Gene.
2021;
30
:
100976
.
View Article Google Scholar -
Li
S.,
Zhao
L.,
Wan
X.H.,
A missense variant rs4645843 in TNF-α gene is a risk factor of polycystic ovary syndrome in the Uygur population. The Tohoku Journal of Experimental Medicine.
2017;
243
(2)
:
95-100
.
View Article PubMed Google Scholar -
Zhang
Y.,
Che
L.,
Zhang
M.,
He
J.,
Common cytokine polymorphisms and predisposition to polycystic ovary syndrome: a meta-analysis. Endocrine Journal.
2020;
67
(5)
:
561-7
.
View Article PubMed Google Scholar -
Mao
Z.,
Fan
L.,
Yu
Q.,
Luo
S.,
Wu
X.,
Tang
J.,
Abnormality of klotho signaling is involved in polycystic ovary syndrome. Reproductive Sciences (Thousand Oaks, Calif.).
2018;
25
(3)
:
372-83
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 10 No 10 (2023)
Page No.: 5972-5986
Published on: 2023-10-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 4305 times
- PDF downloaded - 1315 times
- XML downloaded - 107 times
 Biomedpress
Biomedpress















