Abstract
COVID-19 is a global pandemic caused by severe acute respiratory syndrome (SARS) coronavirus- 2 (SARS-CoV-2). The three main receptors used by SARS-CoV-2 to bind and gain entry into human cells are ACE, TMPRSS2, and CD147. These molecular factors have crucial roles in human metabolism and homeostasis, but the upregulation of these factors causes severe diseases such as myocarditis, prostate cancer, and other endocrine-related cancers. Studies have found that once humans come into contact with SARS-CoV-2, the chances of being affected by such disorders increase; indeed, infection with the virus is associated with increased morbidity and mortality from heart attacks and pulmonary inflammation. Notably, exposure to some pesticides, such as chlorpyrifos, cypermethrin, and imidacloprid, which are identified as potential endocrine disruptors, causes such disorders by interfering with hormonal signaling pathways, such as the insulinglucagon pathway and the thyroid pathway. This review focuses on the potential role of pesticides in exacerbating the comorbidities linked with SARS-CoV-2 and their effect on the molecular factors associated with SARS-CoV-2. Understanding the potential therapeutic implications of this link between SARS-CoV-2 severity and pesticides requires further clinical trials and investigations.
Introduction
The year 2019 witnessed one of the major catastrophes of the world. Spreading globally, the novel coronavirus has become one of the greatest threats to humankind. This coronavirus belongs to the severe acute respiratory syndrome coronavirus (SARS-CoV) family1. First identified following an outbreak in 20042 in Yunnan, China, SARS is responsible for multiple respiratory diseases such as the common cold, bronchitis, and pneumonia caused by severe acute respiratory syndrome coronavirus (SARS-CoV), which also caused the viral pandemic outbreak in 2019 (COVID-19)3. Owing to the high nucleotide substitution and recombination capacity of coronaviruses, SARS-CoV-2 is still mutating and evolving rapidly4, 2. The severe outbreak of the novel strain SARS-CoV-2 in December 2019 in Wuhan Province of China caused a global breakdown in the economy while still posing a threat to human health, leading to unpredictable disastrous consequences5. SARS-CoVs are generally enveloped viruses with a single-stranded RNA genome6. The International Committee for Taxonomy of Viruses has classified the human coronavirus under the Coronaviridae family, which is genotypically and serologically further divided into four major genera: alpha-CoV, beta-CoV, gamma-CoV, and delta-CoV2. Before the emergence of human coronavirus, there was evidence for the presence and harboring of coronavirus in various animals, such as bovine coronavirus (BCoV) and feline infectious peritonitis virus (FIPV)7. Over time, the parallel development of poultry farming and urbanization, and the frequent close contact of animals with one another and their exposure to humans contributed to the evolution and transmission of coronavirus in other species, such as bats8, 9. Direct contact of humans with these coronavirus-harboring bats in 2019 has been suggested as a possible mode of transmission of the virus in humans10. Since then, SARS-CoV, which causes COVID-19, has affected many countries globally11. SARS-CoV-2 relies on ACE2, TMPRSS2, and CD147 to gain entry into human cells (Figure 1)12. ACE2, TMPRSS2, and CD147 receptors are present on the cells of various organs13 and, by attaching itself to these receptors through various mechanisms, SARS-CoV-2 is able to enter various organs of the human body14.
SARS-CoV-2 is composed of a phosphorylated nucleocapsid (N) protein containing genomic RNA5. This core unit is packed by a phospholipid bilayer envelope to form particles varying in shape and size ranging between 80-120 nm15. The characteristic feature of this assembly is characterized by sharp projections known as spike or spike proteins (S) present on the outer surface16. Regarding its protein composition, SARS-CoV-2 is composed of four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins1. Only three viral proteins, S, E, and M, are embedded in the viral envelope, while the N protein is located in the core of the virus bound to the viral genomic RNA17. The S protein is a glycoprotein assembled as a trimeric unit and mediates receptor binding and membrane fusion, allowing the penetration and entry of the virus into host cells18. The genetic composition of SARS-CoV-2 is nonsegmented plus-sense single-stranded 26-31 kb RNA with varied G+C content (32%–43%)19. The genome composition is: 5′-leader-UTR-replicase/transcriptase-spike (S)-envelope (E)-membrane (M)-nucleocapsid (N)-3′UTR-poly (A) tail20. The 5′UTR and 3′UTR are involved in RNA-RNA interactions during the binding of viruses with other cellular proteins21.
The virus assembly consists of 4 structural proteins: nucleocapsid (N), envelope (E), spike (S), and membrane (M) protein1. After the synthesis of the SARS-CoV-2 structural proteins and genomic RNA at the replication site, an unknown mechanism translocates these entities at the ER-Golgi intermediate compartment (ERGIC) for the assembly of the virus and budding22, during which the structural proteins become embedded in the outer structure, while the N protein packs itself along with the genomic RNA, making up the virion23. The E and M proteins of the virus assembly help in budding, whereas the S protein is involved in the initial host-virus interaction24. The first step in the interaction is attachment of the virus to the cell membrane of the host, which is regulated by the S glycoprotein25. The S protein organizes itself in an identical trimeric manner23. Multiple copies of this structure are embedded in the viral envelope membrane, and this glycoprotein moiety is recognized by host cell receptors26. During biosynthesis and maturation in the infected cell, the recognition phase occurs, at which point this trimeric unit is usually cleaved from the viral structure by the protease furin from the Golgi apparatus27, leaving only two subunits behind, i.e., S1 and S2. The S1 subunit is recognized by angiotensin-converting enzyme (ACE2) and binds to it1. After budding, the virus moves to the ERGIC lumen and reaches the plasma membrane, traveling through secretory pathways where the S2 subunit fuses with the membrane mediated by a fusion peptide (FP)28.
Involvement of SARS-CoV-2 - associated molecular factors
ACE2
Angiotensin-converting enzyme 2 (ACE2) is an enzyme found in various locations in the human body29. It is mostly found attached to the cell membranes of the heart, intestines, gall bladder, testes, and kidney30, 31. It exists in two forms: membrane-bound (mACE2) and soluble (sACE2)32, which make up the renin-angiotensin-aldosterone system (RAAS) that maintains the body’s blood pressure33. mACE2 acts on the enzyme ADAM17, cleaving its extracellular domain to create sACE2, which catalyzes the hydrolysis of angiotensin II into angiotensin34. Angiotensin II is a vasoconstrictor peptide angiotensin (1-7)35 and induces vasodilation after binding with MasR receptors, hence lowering blood pressure and antagonizing the effects of ACE236. This phenomenon is a potential target for drugs that are used in cardiovascular disease treatment37. ACE2 (membrane-bound) is also functionally known as the receptor for the spike glycoprotein of the human coronavirus SARS-CoV-238. Recognizing this interaction of ACE2 with SARS-CoV-2 has accelerated some novel therapeutic approaches to reduce ACE2 expression or block the enzyme to prevent the cellular entry of SARS-CoV-2 in the kidney, heart, lung, and brain, where ACE2 is expressed39.
TMPRSS2
Transmembrane serine protease 2 (TMPRSS2) is a cell surface protein located in endothelial cells lining the heart, liver, respiratory, and digestive tracts40. As a serine protease, it is functionally involved in the cleavage of peptide bonds of proteins that have serine as the nucleophilic amino acid within the active site41. It is encoded by the TMPRSS2 gene and is a member of the TMPRSS family of transmembrane proteins exhibiting serine protease activity42. The protease activity of this protein is used by SARS-CoV-2 to enter cells43. Mutations of the TMPRSS2 gene can lead to prostate cancer. The overexpression of TMPRSS2 and prostate carcinogenesis involves the interplay of the transcription factors ERG and ETV144, 45. Their enhanced expression stimulates the downregulation of androgen receptor signaling46. The protease activity of TMPRSS2 is exploited by SARS-CoV-2 to gain entry into the heart, liver, and respiratory cells40. The S1 and S2 subunits of the virus are cleaved by the proteolytic activity of TMPRSS229.
CD147
In addition to ACE2 and TMPRSS2, another receptor, CD147, is also responsible for SARS-CoV-2 entry into the epithelial cell lining12. CD147 is a transmembrane receptor that forms a transmembrane supramolecular complex and interacts with several extracellular and intracellular factors47. CD147 suppresses NOD2 and the gamma‐secretase protein complex48. NOD2 is part of the innate immune system, and gamma-secretase is responsible for the cleavage of beta-amyloid precursor from the plasma membrane17. Owing to its diverse interactions, CD147 has crucial roles in cell metabolism, motility, and activation49. The initiation of programmed cell death, lymphopenia, and cellular overactivation is suggested to occur as a result of the CD147-SARS-CoV-2 interaction50. Cyclophilins A and B are the two main extracellular ligands of CD147 that are recognized by the nucleocapsid protein and then bind with the spike protein of SARS-CoV-249. The presence of three Asn glycosylation sites in this receptor is also suggested to bind with the glycosylated spike protein of SARS-CoV-251, 52.
Effect of pesticide exposure on ACE2, TMPRSS2 and CD147
Endocrine disruptors (EDs) are chemicals that disturb normal human body metabolism by interfering with hormones in the endocrine system53. These chemicals are known to cause developmental, neurological, reproductive, and immunological disorders54 and are present in plasticware, detergents, cosmetics, and pesticides55. This review focuses on pesticides as endocrine disruptors. Human exposure to such chemicals can cause cardiometabolic diseases, hypertension, diabetes, and other endocrine-related cancers56. With the expansion of SARS-CoV-2 worldwide, the risk of severe diseases such as asthma, cancer, cardiovascular disease, hypertension, diabetes57, and obesity also increased, suggesting a possible connection of these chemicals with SARS-CoV-2.
Although these relationships have not yet been explored to any great extent, several studies hint at the possible involvement of ED chemicals in SARS-CoV-2 susceptibility. Pesticides such as chlorpyrifos, deltamethrin, cypermethrin, and imidacloprid have been identified as possible endocrine disruptors58. Using computational approaches, some of the pathways have been identified as potential targets of endocrine disruptors that contribute to COVID-19 severity59. These signaling pathways include TNF60, insulin resistance, endocrine resistance, MAPK61, IL-17, and prolactin pathways62. Ivermectin is an insecticide that has been identified to show mosquitocidal effects. A study by Lehrer et al. showed the interaction of ivermectin binding with the SARS-CoV-2 spike-RBD-ACE2 complex63. A study of cotton aphids exposed to omethoate, an insecticide, was found to downregulate ACE2 mRNA64. The insecticide methamidophos has been shown to affect ACE2 levels in Oomyzus sokolowskii 65, while some organophosphates have been found to affect ACE2 via RNA interference (RNAi)66.
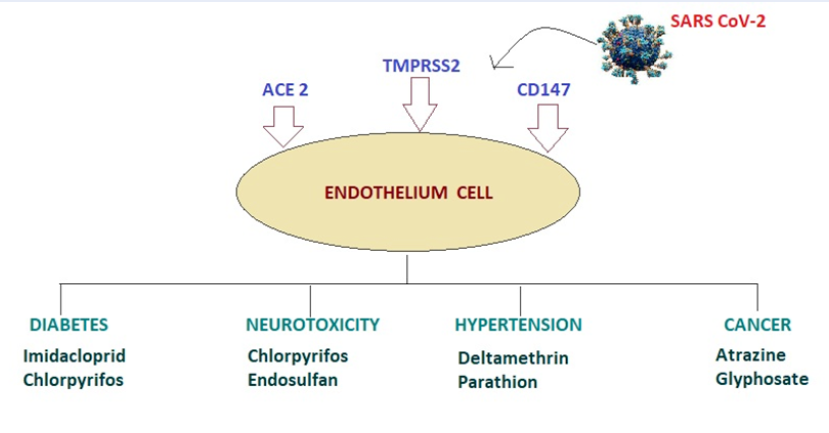
Pesticides are a broad range of chemicals widely used in domestic settings and agriculture67. Exposure to pesticides such as organochlorines, pyrethroids, organophosphates, and carbamates results in disturbances in the normal hormonal functioning of the human body68. Therefore, these chemicals are also classified as endocrine disruptors67. Exposure to these chemicals with human mitochondrial metabolic factors such as ACE2, TMRPSS2, and CD147 also opens the door for SARS-CoV-2 involvement69 (Figure 2). The novel beta-SARS-CoV-2 caused the global health crisis COVID-192. When the virus comes into contact with its living host, it targets ACE2, TMPRSS2, and CD14770. Environmental pollutants can have a wide range of effects on human metabolism. It has been reported that particulate matter from smoking or factories can increase SARS-CoV-2 severity71, but it is unclear whether these pesticides can make organs more susceptible to SARS-CoV-272. Studies have revealed that activation of the IL-8/CXCR1/2 pathway in lung fibroblasts is triggered by pesticides and air pollutants62, resulting in upregulation of ACE2 and TMPRSS2 levels, thereby increasing the risk of SARS-CoV-2 infection. The upregulation was prevented by blocking the IL-8/CXCR1/2 pathway73. Clinical trials are required to determine whether pesticides are linked with SARS-CoV-2 severity and, if yes, it is important to directly or indirectly uncover the underlying mechanisms and pathways74. Given the importance of TMPRSS2 in SARS-CoV-2 infection, a number of studies have considered potential therapeutic approaches that might target this receptor. Certain selective bioactive compounds, from organisms such as P. grandiflorus, show promise as an alternative treatment option against SARS-CoV-2 infections by targeting TMPRSS275. Similarly, designing peptide-mimicking compounds that can inhibit TMPRSS2 activity is another possible approach76. α-Terthienyl, a new-generation insecticide, downregulates TMPRSS2 by reducing its mRNA and protein expression77 but increases the expression of p27, a tumor suppressor gene78. Some anti-CD147 drugs, such as meplazumab, inhibit SARS-CoV-2 in patients with COVID‐19 pneumonia79. Another approach is the truncation of the cytoplasmic tail of CD147 to prevent the entry of SARS‐CoV‐280. As discussed, some environmental toxicants (pesticides) are known to modulate ACE2, TMPRSS2, and CD147 expression thus, further preclinical studies and clinical trials are required to assess inhibitory compounds against these toxicants81.
Conclusion
The receptors ACE2, TMPRSS2 and CD147 are the main entry for the SARS-CoV-2 in humans. These molecular factors play a crucial role in human metabolism and homeostatis but their overexpression and upregulation leads to various pathophysiological conditions. The endocrine disrupting pesticides such as chlorpyripos, cypermethrin, imidacloprid modulate the expression of ACE2, TMPRSS2 and CD147. The COVID-19 is associated with severe inflammatory conditions leading to cardiovascular and pulmonary pathologies. Hence, the exposure of these endocrine disrupting pesticides will increase the comorbidities associated with SARS-CoV-2 infection by interfering with the hormonal signaling pathways. Hence, the present review article explores the link between pesticide exposure and endocrine receptors associated with SARS-CoV-2 infection.
Abbreviations
ACE2: angiotensin onverting enzyme 2; CD147: cluster of differentiation 147; COVID-19: coronavirus disease-2019; ED: endocrine disrupting/disruptors; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; TMPRSS2: transmembrane serine protease 2
Acknowledgments
None.
Author’s contributions
SD: data collection; analysis and interpretation of results; draft manuscript preparation. HA: study conception and design; analysis and interpretation of results; draft manuscript preparation; correction and proofreading of manuscript. FHK: study conception and design; analysis and interpretation of results; All authors approved the final version of the manuscript for submission.
Funding
None.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Jackson
C.B.,
Farzan
M.,
Chen
B.,
Choe
H.,
Mechanisms of SARS-CoV-2 entry into cells. Nature Reviews. Molecular Cell Biology.
2022;
23
(1)
:
3-20
.
View Article PubMed Google Scholar -
Wu
F.,
Zhao
S.,
Yu
B.,
Chen
Y.M.,
Wang
W.,
Song
Z.G.,
Hu
Y.,
Tao
Z.W.,
Tian
J.H.,
Pei
Y.Y.,
Yuan
M.L.,
Zhang
Y.L.,
Dai
F.H.,
Liu
Y.,
Wang
Q.M.,
Zheng
J.J.,
Xu
L.,
Holmes
E.C.,
Zhang
Y.Z.,
A new coronavirus associated with human respiratory disease in China. Nature.
2020;
2020
(579)
:
265-580:E7
.
View Article Google Scholar -
Pene
F.,
Merlat
A.,
Vabret
A.,
Rozenberg
F.,
Buzyn
A.,
Dreyfus
F.,
Coronavirus 229E-related pneumonia in immunocompromised patients. Clinical Infectious Diseases.
2003;
37
(7)
:
929-32
.
View Article PubMed Google Scholar -
Vijgen
L.,
Keyaerts
E.,
Moës
E.,
Maes
P.,
Duson
G.,
Van Ranst
M.,
Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. Journal of Clinical Microbiology.
2005;
43
(11)
:
5452-6
.
View Article PubMed Google Scholar -
Zhou
P.,
Yang
X.L.,
Wang
X.G.,
Hu
B.,
Zhang
L.,
Zhang
W.,
A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature.
2020;
579
(7798)
:
270-3
.
View Article PubMed Google Scholar -
Masters
P.S.,
The molecular biology of coronaviruses. Advances in Virus Research.
2006;
66
:
193-292
.
View Article PubMed Google Scholar -
Saif
L.J.,
Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome?. Revue Scientifique et Technique (International Office of Epizootics).
2004;
23
(2)
:
643-60
.
View Article PubMed Google Scholar -
Xie
J.,
Li
Y.,
Shen
X.,
Goh
G.,
Zhu
Y.,
Cui
J.,
Dampened STING-dependent interferon activation in bats. Cell Host & Microbe.
2018;
23
(3)
:
P297-301.e4
.
View Article PubMed Google Scholar -
Gouilh
M. Ar,
Puechmaille
S.J.,
Diancourt
L.,
Vandenbogaert
M.,
Serra-Cobo
J.,
Roig
M. Lopez,
consortium
EPICOREM,
SARS-CoV related Betacoronavirus and diverse Alphacoronavirus members found in western old-world. Virology.
2018;
517
:
88-97
.
View Article PubMed Google Scholar -
Banerjee
A.,
Kulcsar
K.,
Misra
V.,
Frieman
M.,
Mossman
K.,
Bats and Coronaviruses. Viruses.
2019;
11
(1)
:
41
.
View Article PubMed Google Scholar -
Kirtipal
N.,
Bharadwaj
S.,
Kang
S.G.,
From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infection, Genetics and Evolution.
2020;
85
:
104502
.
View Article PubMed Google Scholar -
Wang
K.,
Chen
W.,
Zhou
Y.S.,
SARS‐CoV‐2 invades host cells via a novel route: CD147‐spike protein. bioRxiv. 2020:2020.2003.2014.988345..
2020
.
View Article Google Scholar -
Lan
J.,
Ge
J.,
Yu
J.,
Shan
S.,
Zhou
H.,
Fan
S.,
Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature.
2020;
581
(7807)
:
215-20
.
View Article PubMed Google Scholar -
Walls
A.C.,
Park
Y.J.,
Tortorici
M.A.,
Wall
A.,
McGuire
A.T.,
Veesler
D.,
Structure, function, and antigenicity of the SARS- CoV-2 spike glycoprotein. Cell.
2020;
181
(2)
.
View Article PubMed Google Scholar -
Bárcena
M.,
Oostergetel
G.T.,
Bartelink
W.,
Faas
F.G.,
Verkleij
A.,
Rottier
P.J.,
Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proceedings of the National Academy of Sciences of the United States of America.
2009;
106
(2)
:
582-7
.
View Article PubMed Google Scholar -
Neuman
B.W.,
Adair
B.D.,
Yoshioka
C.,
Quispe
J.D.,
Orca
G.,
Kuhn
P.,
Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. Journal of Virology.
2006;
80
(16)
:
7918-28
.
View Article PubMed Google Scholar -
Zhou
S.,
Zhou
H.,
Walian
P.J.,
Jap
B.K.,
CD147 is a regulatory subunit of the gamma-secretase complex in Alzheimer's disease amyloid beta-peptide production. Proceedings of the National Academy of Sciences of the United States of America.
2005;
102
(21)
:
7499-504
.
View Article PubMed Google Scholar -
Bosch
B.J.,
van der Zee
R.,
de Haan
C.A.,
Rottier
P.J.,
The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. Journal of Virology.
2003;
77
(16)
:
8801-11
.
View Article PubMed Google Scholar -
Yang
H.,
Bartlam
M.,
Rao
Z.,
Drug design targeting the main protease, the Achilles' heel of coronaviruses. Current Pharmaceutical Design.
2006;
12
(35)
:
4573-90
.
View Article PubMed Google Scholar -
Mousavizadeh
L.,
Ghasemi
S.,
Genotype and phenotype of COVID-19: Their roles in pathogenesis. Journal of Microbiology, Immunology and Infection.
2021;
54
(2)
:
159-63
.
View Article Google Scholar -
Yang
D.,
Leibowitz
J.L.,
The structure and functions of coronavirus genomic 3' and 5' ends. Virus Research.
2015;
206
:
120-33
.
View Article PubMed Google Scholar -
Stertz
S.,
Reichelt
M.,
Spiegel
M.,
Kuri
T.,
Martínez-Sobrido
L.,
García-Sastre
A.,
The intracellular sites of early replication and budding of SARS-coronavirus. Virology.
2007;
361
(2)
:
304-15
.
View Article PubMed Google Scholar -
V'kovski
P.,
Kratzel
A.,
Steiner
S.,
Stalder
H.,
Thiel
V.,
Coronavirus biology and replication: implications for SARS-CoV-2. Nature Reviews. Microbiology.
2021;
19
(3)
:
155-70
.
View Article PubMed Google Scholar -
Goldsmith
C.S.,
Tatti
K.M.,
Ksiazek
T.G.,
Rollin
P.E.,
Comer
J.A.,
Lee
W.W.,
Ultrastructural characterization of SARS coronavirus. Emerging Infectious Diseases.
2004;
10
(2)
:
320-6
.
View Article PubMed Google Scholar -
Shang
J.,
Wan
Y.,
Luo
C.,
Ye
G.,
Geng
Q.,
Auerbach
A.,
Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America.
2020;
117
(21)
:
11727-34
.
View Article PubMed Google Scholar -
Schoeman
D.,
Fielding
B.C.,
Coronavirus envelope protein: current knowledge. Virology Journal.
2019;
16
(1)
:
69
.
View Article PubMed Google Scholar -
Hoffmann
M.,
Kleine-Weber
H.,
Pöhlmann
S.,
Multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Molecular Cell.
2020;
78
(4)
.
View Article PubMed Google Scholar -
Fehr
A.R.,
Perlman
S.,
Coronaviruses: an overview of their replication and pathogenesis. Methods in Molecular Biology (Clifton, N.J.).
2015;
1282
:
1-23
.
View Article PubMed Google Scholar -
NCBI "Gene: ACE2, angiotensin I converting enzyme 2". National Center for Biotechnology Information (NCBI). U.S. National Library of Medicine. 2020-02-28. 2020
.
-
Hikmet
F.,
Méar
L.,
Edvinsson
A.A.,
Micke
P.,
Uhlén
M.,
Lindskog
C.,
The protein expression profile of ACE2 in human tissues. Molecular Systems Biology.
2020;
16
(7)
:
e9610
.
View Article PubMed Google Scholar -
Hamming
I.,
Timens
W.,
Bulthuis
M.L.,
Lely
A.T.,
Navis
G.,
van Goor
H.,
Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology.
2004;
203
(2)
:
631-7
.
View Article PubMed Google Scholar -
Diz
D.I.,
Garcia-Espinosa
M.A.,
Gegick
S.,
Tommasi
E.N.,
Ferrario
C.M.,
Ann Tallant
E.,
Injections of angiotensin-converting enzyme 2 inhibitor MLN4760 into nucleus tractus solitarii reduce baroreceptor reflex sensitivity for heart rate control in rats. Experimental Physiology.
2008;
93
(5)
:
694-700
.
View Article PubMed Google Scholar -
Dong
B.,
Zhang
C.,
Feng
J.B.,
Zhao
Y.X.,
Li
S.Y.,
Yang
Y.P.,
Overexpression of ACE2 enhances plaque stability in a rabbit model of atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology.
2008;
28
(7)
:
1270-6
.
View Article PubMed Google Scholar -
Ferrario
C.M.,
Jessup
J.,
Gallagher
P.E.,
Averill
D.B.,
Brosnihan
K.B.,
Ann Tallant
E.,
Effects of renin-angiotensin system blockade on renal angiotensin-(1-7) forming enzymes and receptors. Kidney International.
2005;
68
(5)
:
2189-96
.
View Article PubMed Google Scholar -
Hoffmann
M.,
Kleine-Wever
H.,
Kruger
N.,
Muller
M.,
Drotsten
C.,
Pholhlmann
S.,
The novel coronavirus 2019 (2019-nCoV) uses the SARS coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry in target cells. Cell.
2020;
181
:
1-10
.
-
Garabelli
P.J.,
Modrall
J.G.,
Penninger
J.M.,
Ferrario
C.M.,
Chappell
M.C.,
Distinct roles for angiotensin-converting enzyme 2 and carboxypeptidase A in the processing of angiotensins within the murine heart. Experimental Physiology.
2008;
93
(5)
:
613-21
.
View Article PubMed Google Scholar -
Jia
H.P.,
Look
D.C.,
Tan
P.,
Shi
L.,
Hickey
M.,
Gakhar
L.,
Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. American Journal of Physiology. Lung Cellular and Molecular Physiology.
2009;
297
(1)
:
84-96
.
View Article PubMed Google Scholar -
Kuba
K.,
Imai
Y.,
Rao
S.,
Gao
H.,
Guo
F.,
Guan
B.,
A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nature Medicine.
2005;
11
(8)
:
875-9
.
View Article PubMed Google Scholar -
Wrapp
D.,
Wang
N.,
Corbett
K.S.,
Goldsmith
J.A.,
Hsieh
C.L.,
Abiona
O.,
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science.
2020;
367
(6483)
:
1260-3
.
View Article PubMed Google Scholar -
Bertram
S.,
Glowacka
I.,
Blazejewska
P.,
Soilleux
E.,
Allen
P.,
Danisch
S.,
TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. Journal of Virology.
2010;
84
(19)
:
10016-25
.
View Article PubMed Google Scholar -
Thunders
M.,
Delahunt
B.,
Gene of the month: TMPRSS2 (transmembrane serine protease 2). Journal of Clinical Pathology.
2020;
73
(12)
:
773-6
.
View Article PubMed Google Scholar -
Gierer
S.,
Bertram
S.,
Kaup
F.,
Wrensch
F.,
Heurich
A.,
Krämer-Kühl
A.,
The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. Journal of Virology.
2013;
87
(10)
:
5502-11
.
View Article PubMed Google Scholar -
Bossart
K.N.,
Wang
L.F.,
Flora
M.N.,
Chua
K.B.,
Lam
S.K.,
Eaton
B.T.,
Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. Journal of Virology.
2002;
76
(22)
:
11186-98
.
View Article PubMed Google Scholar -
Winnes
M.,
Lissbrant
E.,
Damber
J.E.,
Stenman
G.,
Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncology Reports.
2007;
17
(5)
:
1033-6
.
View Article PubMed Google Scholar -
Beuzeboc
P.,
Soulié
M.,
Richaud
P.,
Salomon
L.,
Staerman
F.,
Peyromaure
M.,
[Fusion genes and prostate cancer. From discovery to prognosis and therapeutic perspectives]. Progres en Urologie.
2009;
19
(11)
:
819-24
.
View Article PubMed Google Scholar -
Yu
J.,
Yu
J.,
Mani
R.S.,
Cao
Q.,
Brenner
C.J.,
Cao
X.,
An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell.
2010;
17
(5)
:
443-54
.
View Article PubMed Google Scholar -
Muramatsu
T.,
Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. Journal of Biochemistry.
2016;
159
(5)
:
481-90
.
View Article PubMed Google Scholar -
Till
A.,
Rosenstiel
P.,
Bräutigam
K.,
Sina
C.,
Jacobs
G.,
Oberg
H.H.,
A role for membrane-bound CD147 in NOD2-mediated recognition of bacterial cytoinvasion. Journal of Cell Science.
2008;
121
(Pt 4)
:
487-95
.
View Article PubMed Google Scholar -
Radzikowska
U.,
Ding
M.,
Tan
G.,
Zhakparov
D.,
Peng
Y.,
Wawrzyniak
P.,
Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy.
2020;
75
(11)
:
2829-45
.
View Article PubMed Google Scholar -
Goyal
P.,
Choi
J.J.,
Pinheiro
L.C.,
Schenck
E.J.,
Chen
R.,
Jabri
A.,
Clinical Characteristics of Covid-19 in New York City. The New England Journal of Medicine.
2020;
382
(24)
:
2372-4
.
View Article PubMed Google Scholar -
Huang
W.,
Luo
W.J.,
Zhu
P.,
Tang
J.,
Yu
X.L.,
Cui
H.Y.,
Modulation of CD147-induced matrix metalloproteinase activity: role of CD147 N-glycosylation. The Biochemical Journal.
2013;
449
(2)
:
437-48
.
View Article PubMed Google Scholar -
Vankadari
N.,
Wilce
J.A.,
Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerging Microbes & Infections.
2020;
9
(1)
:
601-4
.
View Article PubMed Google Scholar -
Dixit
S.,
Zia
M.K.,
Siddiqui
T.,
Ahsan
H.,
Khan
F.H.,
Interaction of Human Alpha-2-Macroglobulin with Pesticide Aldicarb Using Spectroscopy and Molecular Docking. Protein and Peptide Letters.
2021;
28
(3)
:
315-22
.
View Article PubMed Google Scholar -
Mnif
W.,
Hassine
A.I.,
Bouaziz
A.,
Bartegi
A.,
Thomas
O.,
Roig
B.,
Effect of endocrine disruptor pesticides: a review. International Journal of Environmental Research and Public Health.
2011;
8
(6)
:
2265-303
.
View Article PubMed Google Scholar -
Darbre
P.D.,
Chemical components of plastics as endocrine disruptors: overview and commentary. Birth Defects Research.
2020;
112
(17)
:
1300-7
.
View Article PubMed Google Scholar -
Zahra
A.,
Sisu
C.,
Silva
E.,
De Aguiar Greca
S.C.,
Randeva
H.S.,
Chatha
K.,
Is There a Link between Bisphenol A (BPA), a Key Endocrine Disruptor, and the Risk for SARS-CoV-2 Infection and Severe COVID-19?. Journal of Clinical Medicine.
2020;
9
(10)
:
3296
.
View Article PubMed Google Scholar -
Sandoval
M.,
Nguyen
D.T.,
Vahidy
F.S.,
Graviss
E.A.,
Risk factors for severity of COVID-19 in hospital patients age 18-29 years. PLoS One.
2021;
16
(7)
:
e0255544
.
View Article PubMed Google Scholar -
Abdel-Razik
R.K.,
Mosallam
E.M.,
Hamed
N.A.,
Badawy
M.E.,
Abo-El-Saad
M.M.,
Testicular deficiency associated with exposure to cypermethrin, imidacloprid, and chlorpyrifos in adult rats. Environmental Toxicology and Pharmacology.
2021;
87
:
103724
.
View Article PubMed Google Scholar -
Wu
Q.,
Coumoul
X.,
Grandjean
P.,
Barouki
R.,
Audouze
K.,
Endocrine disrupting chemicals and COVID-19 relationships: a computational systems biology approach. medRxiv.
2020;
2020
.
View Article Google Scholar -
Fabregat
A.,
Jupe
S.,
Matthews
L.,
Sidiropoulos
K.,
Gillespie
M.,
Garapati
P.,
The Reactome Pathway Knowledgebase. Nucleic Acids Research.
2018;
46
:
649-55
.
View Article PubMed Google Scholar -
Kanehisa
M.,
Sato
Y.,
Furumichi
M.,
Morishima
K.,
Tanabe
M.,
New approach for understanding genome variations in KEGG. Nucleic Acids Research.
2019;
47
:
590-5
.
View Article PubMed Google Scholar -
Slenter
D.N.,
Kutmon
M.,
Hanspers
K.,
Riutta
A.,
Windsor
J.,
Nunes
N.,
WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Research.
2018;
46
:
661-7
.
View Article PubMed Google Scholar -
Lehrer
S.,
Rheinstein
P.H.,
Ivermectin Docks to the SARS-CoV-2 Spike Receptor-binding Domain Attached to ACE2. In Vivo (Athens, Greece).
2020;
34
(5)
:
3023-6
.
View Article PubMed Google Scholar -
Pan
Y.,
Shang
Q.,
Fang
K.,
Zhang
J.,
Xi
J.,
Down-regulated transcriptional level of Ace1 combined with mutations in Ace1 and Ace2 of Aphis gossypii are related with omethoate resistance. Chemico-Biological Interactions.
2010;
188
(3)
:
553-7
.
View Article PubMed Google Scholar -
Zhuang
H.M.,
Li
C.W.,
Wu
G.,
Identification and characterization of ace2-type acetylcholinesterase in insecticide-resistant and -susceptible parasitoid wasp Oomyzus sokolowskii (Hymenoptera: eulophidae). Molecular Biology Reports.
2014;
41
(11)
:
7525-34
.
View Article PubMed Google Scholar -
Revuelta
L.,
Piulachs
M.D.,
Bellés
X.,
Castañera
P.,
Ortego
F.,
Díaz-Ruíz
J.R.,
RNAi of ace1 and ace2 in Blattella germanica reveals their differential contribution to acetylcholinesterase activity and sensitivity to insecticides. Insect Biochemistry and Molecular Biology.
2009;
39
(12)
:
913-9
.
View Article PubMed Google Scholar -
Dixit
S.,
Ahsan
H.,
Khan
F.H.,
Pesticides and plasma proteins: unexplored dimensions in neurotoxicity. International Journal of Pest Management.
2021;
69
(3)
:
278-87
.
View Article Google Scholar -
Mi
H.,
Muruganujan
A.,
Thomas
P.D.,
PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research.
2013;
41
(Database issue)
:
377-86
.
PubMed Google Scholar -
Yao
Y.,
Lawrence
D.A.,
Susceptibility to COVID-19 in populations with health disparities: posited involvement of mitochondrial disorder, socioeconomic stress, and pollutants. Journal of Biochemical and Molecular Toxicology.
2021;
35
(1)
:
e22626
.
View Article PubMed Google Scholar -
Duru
C.E.,
Umar
H.I.,
Duru
I.A.,
Enenebeaku
U.E.,
Ngozi-Olehi
L.C.,
Enyoh
C.E.,
Blocking the interactions between human ACE2 and coronavirus spike glycoprotein by selected drugs: a computational perspective. Environmental Analysis, Health and Toxicology.
2021;
36
(2)
:
e2021010
.
View Article PubMed Google Scholar -
Baraniuk
C.,
Receptors for SARS-CoV-2 Present in Wide Variety of Human Cells. Scientist (Philadelphia, Pa.).
2020
.
-
Sagawa
T.,
Tsujikawa
T.,
Honda
A.,
Miyasaka
N.,
Tanaka
M.,
Kida
T.,
Exposure to particulate matter upregulates ACE2 and TMPRSS2 expression in the murine lung. Environmental Research.
2021;
195
:
110722
.
View Article PubMed Google Scholar -
Li
H.H.,
Liu
C.C.,
Hsu
T.W.,
Lin
J.H.,
Hsu
J.W.,
Li
A.F.,
Upregulation of ACE2 and TMPRSS2 by particulate matter and idiopathic pulmonary fibrosis: a potential role in severe COVID-19. Particle and Fibre Toxicology.
2021;
18
(1)
:
11
.
View Article PubMed Google Scholar -
South
A.M.,
Diz
D.I.,
Chappell
M.C.,
COVID-19, ACE2, and the cardiovascular consequences. American Journal of Physiology. Heart and Circulatory Physiology.
2020;
318
(5)
:
1084-90
.
View Article PubMed Google Scholar -
Gurung
A.B.,
Ali
M.A.,
Lee
J.,
Aljowaie
R.M.,
Almutairi
S.M.,
Exploring the phytochemicals of Platycodon grandiflorus for TMPRSS2 inhibition in the search for SARS-CoV-2 entry inhibitors. Journal of King Saud University. Science.
2022;
34
(6)
:
102155
.
View Article PubMed Google Scholar -
Shapira
T.,
Monreal
I.A.,
Dion
S.P.,
Buchholz
D.W.,
Imbiakha
B.,
Olmstead
A.D.,
TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature.
2022;
605
(7909)
:
340-8
.
View Article PubMed Google Scholar -
Mahoney
M.,
Damalanka
V.C.,
Tartell
M.A.,
Chung
D.H.,
Lourenço
A.L.,
Pwee
D.,
A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proceedings of the National Academy of Sciences of the United States of America.
2021;
118
(43)
:
e2108728118
.
View Article PubMed Google Scholar -
Gan
X.,
Huang
H.,
Wen
J.,
Liu
K.,
Yang
Y.,
Li
X.,
α-Terthienyl induces prostate cancer cell death through inhibiting androgen receptor expression. Biomedicine and Pharmacotherapy.
2022;
152
:
113266
.
View Article PubMed Google Scholar -
Bian
H.,
Zheng
Z.H.,
Wei
D.,
Meplazumab treats COVID‐19 pneumonia: an open‐labelled, concurrent controlled add‐on clinical trial. medRxiv. 2020:2020.2003.2021.20040691.. 2020
.
View Article Google Scholar -
Pushkarsky
T.,
Yurchenko
V.,
Laborico
A.,
Bukrinsky
M.,
CD147 stimulates HIV-1 infection in a signal-independent fashion. Biochemical and Biophysical Research Communications.
2007;
363
(3)
:
495-9
.
View Article PubMed Google Scholar -
Rath
S.,
Perikala
V.,
Jena
A.B.,
Dandapat
J.,
Factors regulating dynamics of angiotensin-converting enzyme-2 (ACE2), the gateway of SARS-CoV-2: epigenetic modifications and therapeutic interventions by epidrugs. Biomedicine and Pharmacotherapy.
2021;
143
:
112095
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 10 No 9 (2023)
Page No.: 5876-5883
Published on: 2023-09-30
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 4462 times
- PDF downloaded - 1188 times
- XML downloaded - 92 times
 Biomedpress
Biomedpress



