Abstract
Introduction: Human exfoliated deciduous teeth?derived stem cells (SHEDs) have been shown as an excellent source of bone regeneration. Interleukin-17A (IL-17A) facilitates bone differentiation in various cell types, including SHEDs. In this study, we have demonstrated IL-17A?s stimulating effect on SHEDs in osteogenic differentiation and further evaluated the role of the ERK/MAPK signaling pathway in this process.
Methods: The function of IL-17A in osteogenic differentiation, proliferative activity, and MAPK cascade activation in SHEDs were investigated.
Results: IL-17A significantly enhanced proliferative and alkaline phosphatase activities in SHEDs. Furthermore, the expression levels of different osteogenic proteins including COL1A1, ALP, OPN, RUNX, and OCN were significantly elevated in IL-17A-treated SHEDs. Moreover, IL-17A triggered MAPK signaling in SHEDs, as evidenced by significant upregulation of both downstream ERK targets, P38 and JNK pathways, and upstream activators. In addition, ERK/MAPK activation time-dependently established the participation of MAPK signaling in SHED osteogenic differentiation.
Conclusion: These findings suggest that IL-17A-induced ERK/MAPK signaling pathway activation is necessary for SHEDs to differentiate into osteoblasts. This reiterates the significance of this particular intracellular signaling pathway in controlling SHED osteogenic differentiation, which is a promising source of bone tissue regeneration.
Introduction
The regeneration of bone fractures resulting from trauma, tumors, infection, and degenerative and inflammatory conditions involves a complex physiological process. It includes a series of biological events involving multiple growth factors and cell types, as well as intracellular and extracellular molecular-signaling pathways1. Tight interactions between the immune system and the regeneration process have been well-documented. The proinflammatory response following bone injury is a crucial initiator of bone regeneration2.
Interleukin-17A (IL-17A) is a proinflammatory cytokine produced by a specialized CD4+ T cell subset, T helper 17 (Th17) cells. IL-17A induces signaling by targeting the IL-17A receptor/IL-17C receptor (IL17RA/IL17RC) complex3. IL17RA/IL17RC complexes are expressed not only in the immune cells but also by the non-immune cells and tissues4. The interaction of IL-17A and its receptors stimulates downstream regulators and induces cellular responses5. The intracellular IL-17 downstream signaling comprises mitogen-activated protein kinases (MAPKs) and nuclear factor-κB (NF-κB)6, 7. In the past decade, many studies have demonstrated novel roles of IL-17A in bone homeostasis, promoting either bone formation or bone loss mechanisms8. IL-17A is traditionally recognized as a bone-destructive cytokine in different immune-related bone diseases such as rheumatoid arthritis, osteoporosis, periodontitis, psoriatic arthritis9, and spondylarthritis10. Several studies have shown that IL-17A promotes osteoblast differentiation by upregulating osteogenic genes such as osteoprotegerin (OPG), Runx2, bone sialoprotein (BSP), osteopontin (OPN), osteocalcin and type I collagen (Colla1), and ALP 11, 12, 13. Other studies have revealed that IL−17A suppressed the differentiation of osteoblasts by downregulating bone markers14, 15. IL-17A induced bone marrow–derived mesenchymal stem cells (BM-MSCs) to produce IL-1 and IL-6, which stimulated the STAT3, ERK1/2, and AKT pathways, promoting osteoblastic differentiation16. Moreover, IL-17A facilitates osteogenesis on BM-MSCs by increasing leptin production via JAK/STAT signaling17. Multiple studies have implicated IL-17 as crucial in bone remodeling, with the opposite roles of either enhancing or aggravating osteogenesis based on the microenvironment. However, the precise IL-17A mechanism in the multipotent stem cells from human exfoliated deciduous teeth (SHEDs) remains unclear.
Because of their multipotency and relative ease of accessibility, SHEDs have attracted great attention recently. SHEDs can rapidly regenerate and grow faster than adult stem cells and restore tissue functions through a process known as plasticity or trans-differentiation18. Moreover, SHEDs, which are derived from bone marrow, possess the capacity for osteoblastic differentiation19. In this study, we investigated the stimulatory effects of IL-17A and further elucidated the participation of the MAPK signaling pathway in SHED osteogenic differentiation.
Materials and Methods
Cell culture and osteogenic differentiation in vitro
The SHED cell line, obtained from ALLCELLS (Alameda, CA, USA), was derived from human deciduous teeth of the third molar, which were exfoliated from 7- to 8-year-old children. The cells were cultured in α-minimum essential medium (αMEM; Gibco, Waltham, MA, USA) at 37 °C with 5% CO2 in a humid environment, from passages 3–5. The complete culture medium was prepared using 10% heat-inactivated fetal bovine serum (Gibco), 1% L-glutamine, and 1% penicillin-streptomycin.
For alkaline phosphatase (ALP) and proliferation assays, cells were cultured in complete αMEM medium supplemented with recombinant IL-17A at concentrations of 5, 10, 25, 50, and 100 ng/mL. Proliferation assays were conducted for up to 5 days, and ALP assays were performed at 1, 3, 7, and 14 days.
For osteogenic protein expression and MAPK studies, osteoinducing reagents were added to the complete growth medium to create osteogenic medium (OM), which included 10 mM β-glycerophosphate, 50 µg/mL L-ascorbic acid, and 10 nM dexamethasone. After 24 hours, IL-17A at a concentration of 50 ng/mL (eBiosciences, USA) was added to the cells for treatment, following a previous study20.
SHED characterization
For the characterization process, an inverted microscope (Carl Zeiss, Germany) was employed to observe the cell morphology of SHEDs from passages 3 to 4 on days 1 and 3. Flow cytometry was used to assess the expression of surface markers on SHED cells. Mesenchymal markers were used, including mouse anti-human CD44, mouse anti-human CD105, mouse anti-human c-Myc, and mouse anti-human Nanog. Monoclonal mouse anti-human CD34 was used as a negative marker to characterize SHED cells. Cells were fixed with 4.2% formaldehyde, permeabilized with ice-cold methanol, and blocked with 10% normal goat serum at room temperature. Subsequently, 1 × 106 cells were stained with individual antibodies at a standardized concentration of 1 μg/mL for 1 hour in the dark at room temperature. After two washes with phosphate-buffered saline, secondary antibodies conjugated with goat anti-mouse PE and goat anti-mouse FITC were applied, vortexed carefully, and incubated for 30 minutes in the dark at room temperature. Flow cytometry was performed using a FACSCanto II instrument (Becton, NJ, USA) to assess the cells. Isotype mouse IgG1 was used as a negative control, and human osteoblast cells were used as a positive control.
Cell proliferation
Briefly, the cells were seeded at a density of 2x103 cells/well in 96-well plates. They were then incubated at 37 °C in a humidified atmosphere with 5% CO2 for intervals of 24, 48, 72, 96, and 120 hours. Media changes were conducted every 48 hours. Cell proliferation was measured using the 3‐(4,5‐dimethyl‐thiazol‐2‐yl)‐5‐(3‐carboxy‐methoxy‐phenyl)‐2‐(4‐sulfo‐phenyl)‐2H‐tetrazolium (MTS) assay with CellTiter 96 AQueous One Solution (Promega, Southampton, UK), following the manufacturer’s protocol. The absorbance at 490 nm, indicative of cell viability, was measured using a microplate reader (Sunrise Tecan, Austria) and expressed as optical density (OD). Cell viability was determined as follows:
Percentage of cell viability = [(A-B)/(C-B)] x 100; where A, B, and C are the OD of the sample, blank wells, and control group, respectively.
ALP activity
Alkaline phosphatase (ALP) activity was assessed using the Alkaline Phosphatase Activity Kit from BioVision (MA, USA), following the manufacturer’s protocol. Absorbance at 405 nm was measured using a microplate reader to estimate ALP activity in the experimental samples, and the ALP activity was determined as follows:
ALP activity (U/mL) = A/V/T; where A denotes the pNP amount produced by samples (μmol), V represents the volume of the sample inside the well (mL), and T represents the reaction time (min).
Analysis of bone regulatory protein expression by Western blotting
Western blot analysis was performed to detect collagen type I alpha 1 (COL1A1), alkaline phosphatase (ALP), osteocalcin (OCN), runt-related transcription factor 2 (RUNX2), and osteopontin (OPN) proteins on days 3, 7, and 14. SHEDs were washed and lysed using radioimmunoprecipitation assay (RIPA) buffer, and the protein concentration in the cell lysate was evaluated using a protein assay kit from Bio-Rad (CA, USA). A protein concentration of 20 micrograms per sample was used, which was collected from 3.2 x 106 cells/well in 6-well plates. Protein lysates were separated on 12–15% polyacrylamide gels (PAGs) by electrophoresis and then transferred onto polyvinylidene difluoride (PVDF) membranes. The PVDF membrane was blocked with 5% skimmed milk solution for 1 hour, followed by incubation with purified anti-human monoclonal antibodies against ALP, OCN, COL1A1, RUNX2, and OPN at 4 °C overnight. The membrane was then washed with TBS-T20 and incubated with horseradish peroxidase–conjugated mouse IgG antibody for 1 hour at room temperature. Chemiluminescence was detected using an enhanced chemiluminescence (ECL) Western blot detection reagent, and the integrated density of the bands was calculated using Image J software (Madison, WI, USA) and standardized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Analysis of modulating MAPK signaling using real-time PCR
Real-time PCR (qPCR) was performed using an RT2 Profiler PCR Array from QIAGEN (Hilden, Germany), which included 42 genes related to MAP kinase signaling in subgroups. Total RNA was isolated from IL-17A-treated cells and untreated SHEDs cultured under both OM and without-OM conditions, using the RNeasy Mini Kit (QIAGEN, Germany). Three biological replicates were prepared and harvested on day 7 in accordance with the osteogenic differentiation activity. The amount of RNA (500 ng) was reverse transcribed to cDNA in a final volume of 10 µL by using an RT2 First Strand kit (QIAGEN, Germany) according to the manufacturer’s protocol. qPCR analysis was conducted using RT2 SYBR Green Gene Expression (QIAGEN, Germany) on an ABI Step One Plus Real-time PCR system, and data analysis was performed using the Step One Plus Software version 2.2 Real-Time PCR System from Applied Biosystems, USA. Five housekeeping genes were included in the study: β-actin (ACTB), beta-2 microglobulin (B2M), hypoxanthine phosphoribosyl transferase 1 (HPRT1), GAPDH, and ribosomal protein large P0 (RPLP0). The primers for the 42 genes associated with MAPK, as well as the housekeeping genes, were pre-designed by the manufacturer (QIAGEN, Germany).
To normalize mRNA expression, CT (comparative) values were evaluated during data analysis. The differences in gene expression between the gene of interest and the mean expression of the reference housekeeping genes were calculated using delta–delta CT (delta CT [test group] - delta CT [control group]). This calculation was used to determine the fold-change of each gene. The data were analyzed using the manufacturer’s PCR array data analysis online portal.
Validation of ERK/MAPK expression by Western blot
To detect the expressions of ERK1/2 and p-ERK proteins in IL-17A-treated SHEDs, a Western blot analysis was performed using the described method and reagents. Protein samples were obtained by washing and lysing SHEDs with RIPA buffer. The optimized protein concentration of 20 µg per sample was used in the study. The protein lysates were separated on 12–15% (w/v) polyacrylamide gels and electrophoresed at 90V for 1 hour. Subsequently, the proteins were transferred onto a PVDF membrane, and the membrane was blocked with skimmed milk solution (5%) for 1 hour at room temperature.
Mouse anti-human monoclonal antibodies against ERK1/2, p-ERK, and GAPDH were used for incubation with the PVDF membrane overnight at 4 °C. The membrane was then rinsed with TBS-T20 before being incubated with secondary conjugated-mouse IgG horseradish peroxidase antibody for 1 hour at room temperature. The membrane was then incubated with an ECL Western blot detection reagent to generate chemiluminescence. Protein band integrated density was evaluated using Image J software, and the integrated density of ERK1/2 and p-ERK was normalized to the integrated density of GAPDH, which was used as a loading control.
Statistical analysis
The data from the experiments, which were performed in triplicate (n=3), were analyzed using SPSS software, version 24 (IBM Corp., Armonk, NY, USA). The data are presented as mean ± standard deviation (SD) for individual experiments. For the proliferation assay results, Dunnett T3 post hoc analysis was used for evaluating the statistical significance. One-way ANOVA was performed to analyze the results of Western blot and ALP activity tests, followed by a post hoc multiple comparison test using the Scheffe method. The qPCR data were analyzed using a 2−ΔΔCT approach through web-based software from SABiosciences, USA. The expression level of target genes was normalized to the expression levels of the housekeeping genes. The normality and homogeneity of variance assumptions were checked and fulfilled before performing the statistical analysis. The statistical significance level was set at p < 0.05.
Results
SHED morphology and stem cell marker expression
The SHED morphology was observed on days 1 and 3, as shown in Figure 1A. The SHEDs exhibited a fibroblast-like and spindle-shaped morphology with 1–2 protrusions during culture up to day 3. Further characterization of SHEDs was performed using flow cytometry analysis to assess the expression of stem cell markers. The results showed high expression (more than 95%) of CD105, CD44, Nanog, and c-Myc, which are known markers associated with stem cells. However, low expression of CD34 (less than 2%) was detected, indicating that CD34 expression was relatively low in these SHEDs, as shown in Figure 1B.
IL‐17A increased ALP activity and proliferation in SHED
The results shown in Figure 2A indicate that SHEDs supplemented with varying concentrations of IL-17A exhibited significant (p < 0.05) proliferative activity from days 1 to 5 compared to untreated cells. Specifically, SHEDs supplemented with IL-17A at concentrations of 10, 25, 50, and 100 ng/mL showed significantly higher proliferative activity compared to untreated cells and SHEDs treated with 5 ng/mL IL-17A from day 3 onwards.
In Figure 2B, the ALP activity was evaluated. The results show that ALP activity was enhanced from day 7 until day 14. Specifically, SHEDs supplemented with IL-17A at concentrations of 10, 25, 50, and 100 ng/mL showed significantly higher ALP activity compared to untreated cells and SHEDs treated with 5 ng/mL IL-17A.
These findings suggest that IL-17A supplementation promoted proliferative activity and enhances ALP activity in SHEDs, with higher concentrations of IL-17A showing more significant effects compared to lower concentrations and untreated cells.
IL-17A increased osteogenic protein expressions
The analysis of osteogenic protein expression in SHEDs was conducted by culturing the cells in osteoinducing medium and treating them with 50 ng/mL of IL-17A. Five osteogenic markers associated with bone turnover were assessed in this study: COL1A1, OCN, ALP, OPN, and RUNX2.
Immunoblots (Figure 3A) showed the presence of bands of approximately 60 kDa, 80 kDa, 55 kDa, 6 kDa, and 65 kDa in size, corresponding to COL1A1, OCN, ALP, OPN, and RUNX2 proteins, respectively, in both untreated and treated cells. Untreated cells were used as controls.
Figure 3 B presents the expression levels of COL1A1, OCN, ALP, OPN, and RUNX2 proteins on days 3, 7, and 14 regarding supplementation with 50 ng/mL of IL-17A. The expression of COL1A1 and ALP proteins showed a steady increase on days 3 and 7, reaching their peak expression on day 14. After 2 weeks of stimulation, both COL1A1 and ALP proteins showed significantly elevated expression (p < 0.05) in the IL-17A-treated groups compared to the untreated group. RUNX2, an early osteogenic marker, exhibited the highest expression on day 7. Notably, mature bone differentiation markers such as OPN and OCN showed a gradual increase on days 3 and 7, with the peak expression of proteins observed on day 14 in the IL-17A-supplemented groups (p < 0.05). These findings suggest that IL-17A supplementation promoted the expression of osteogenic markers in SHED, indicating enhanced osteogenic differentiation potential of the cells.
| Gene symbol/ID | Fold change | Gene description | |
|---|---|---|---|
| SHED + IL-17A | SHED + OM + IL-17A | ||
| ERK 1,2 signalling | |||
| EGFR | 2.7 | 14.07 | Epidermal growth factor receptor |
| ARAF | 3.29 | 14.15 | V-raf murine sarcoma 3611 viral oncogene homolog |
| BRAF | 4.35 | 14.44 | V-raf murine sarcoma viral oncogene homolog B1 |
| RAF1 | 5.84 | 19.8 | V-raf-1 murine leukemia viral oncogene homolog 1 |
| KRAS | 4.24 | 13.15 | V-Ki-ras 2 Kirsten rat sarcoma viral oncogene homolog |
| HRAS | 4.2 | 14.11 | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| MAP2K1 | 3.72 | 22.08 | Mitogen-activated protein kinase kinase 1 |
| MAP2K2 | 4.62 | 35.31 | Mitogen-activated protein kinase kinase 2 |
| MAPK1 | 4 | 15.45 | Mitogen-activated protein kinase 1 |
| MAPK3 | 4.71 | 15.66 | Mitogen-activated protein kinase 3 |
| GRB2 | 3.65 | 12.6 | Growth factor receptor-bound protein 2 |
| MOS | 48.16 | 37.99 | V-mos Moloney murine sarcoma viral oncogene homolog |
| MKNK1 | 6.1 | 11.31 | MAP kinase interacting serine/threonine kinase 1 |
| ERK5 Signalling | |||
| MAP3K1 | 3.15 | 38.92 | Mitogen-activated protein kinase kinase kinase 1 |
| MAP3K2 | 3.48 | 13.38 | Mitogen-activated protein kinase kinase kinase 2 |
| MAP3K3 | 5.56 | 13.15 | Mitogen-activated protein kinase kinase kinase 3 |
| MAP2K5 | 3.51 | 14.88 | Mitogen-activated protein kinase kinase 5 |
| MAPK7 | 3.93 | 16.52 | Mitogen-activated protein kinase 7 |
| CREB1 | 4.49 | 13.26 | CAMP responsive element binding protein 1 |
| ERK3 Signalling | |||
| MAPK6 | 5.35 | 14.36 | Mitogen-activated protein kinase 6 |
| p38 Signalling | |||
| MAP4K1 | 32.02 | 55.51 | Mitogen-activated protein kinase kinase kinase kinase 1 |
| MAP2K3 | 4.6 | 19.38 | Mitogen-activated protein kinase kinase 3 |
| MAP2K6 | 3.48 | 8.9 | Mitogen-activated protein kinase kinase 6 |
| MAPK11 | 9.01 | 19.41 | Mitogen-activated protein kinase 11 |
| MAPK12 | 4.26 | 14.53 | Mitogen-activated protein kinase 12 |
| MAPK13 | 2.96 | 8.52 | Mitogen-activated protein kinase 13 |
| MAPK14 | 4.71 | 8.32 | Mitogen-activated protein kinase 14 |
| MAPKAPK2 | 4.25 | 15.61 | Mitogen-activated protein kinase-activated protein kinase 2 |
| MAPKAPK3 | 3.57 | 18.43 | Mitogen-activated protein kinase-activated protein kinase 3 |
| JNK Signalling | |||
| RAC1 | 4.13 | 11.52 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| PAK1 | 4.07 | 13.08 | P21 protein (Cdc42/Rac)-activated kinase 1 |
| MAP3K4 | 4.16 | 13.65 | Mitogen-activated protein kinase kinase kinase 4 |
| MAP2K4 | 5.13 | 14.84 | Mitogen-activated protein kinase kinase 4 |
| MAP2K7 | 2.93 | 22.58 | Mitogen-activated protein kinase kinase 7 |
| MAPK8 | 2.95 | 12.37 | Mitogen-activated protein kinase 8 |
| MAPK9 | 3.32 | 13.22 | Mitogen-activated protein kinase 9 |
| MAPK10 | 6.83 | 12.06 | Mitogen-activated protein kinase 10 |
| Transcription factors common for ERK, p38 and JNK | |||
| ELK1 | 6.45 | 39.35 | Member of ETS oncogene family |
| Transcription factors common for ERK1/2 and ERK5 | |||
| ETS1 | 4.94 | 22.49 | V-etserythroblastosis virus E26 oncogene homolog1 (avian) |
| ETS2 | 4.82 | 21.89 | V-etserythroblastosis virus E26 oncogene homolog2 (avian) |
| Transcription factors common for ERK and p38 | |||
| MYC | 5.7 | 40.92 | V-mycmyelocytomatosis viral oncogene homolog (avian) |
| FOS | 5.46 | 164.29 | FBJ murine osteosarcoma viral oncogene homolog |
| Transcription factors for JNK and p38 | |||
| ATF2 | 4.03 | 13.29 | Activating transcription factor 2 |
| Transcription factors for JNK alone | |||
| JUN | 4.72 | 26.4 | Jun proto-oncogene |
| Osteogenic gene | |||
| COL1A1 | 8.02 | 63.915 | Collagen, type 1, alpha 1 |
IL-17A activated MAP kinase signaling pathway
This study focused on the MKKKK, MAPK, MKKK, and MKK family members and their respective transcription factors, which are modulated through MAP kinase signaling. Figure 4 shows scatter plots of the gene expression in Group 1 (SHED + IL-17A) and Group 2 (SHED + IL-17A + OM). A total of 42 genes were significantly upregulated (p > 0.05) following supplementation with IL-17A in both groups, as shown in Table 1. Interestingly, the mRNA expression of upstream regulators of MAPK signaling pathways was activated during the osteogenic differentiation of SHEDs treated with IL-17A.
Specifically, significant upregulations were observed in the regulators of ERK 1,2 signaling (EGFR, ARAF, BRAF, RAF1, KRAS, MAP2K1, MAP2K2, MAPK1, MAPK3, GRB2, MOS, and MKNK1) in SHEDs treated with IL-17A. Additionally, IL-17A stimulated the activation of some regulators in ERK5 signaling. MAP3K1, MAP3K2, MAP3K3, MAP2K5, MAPK7, and CREB1 were upregulated in both groups treated with IL-17A. In the case of ERK3 signaling, the downstream effector MAPK6 was upregulated. IL-17A also promoted the activation of the p38 signaling pathway, as evidenced by the upregulation of MAP4K1, MAP2K3, MAP2K6, MAPK11, MAPK12, and MAPK13 pathways, along with increased downstream effector kinases MAPKAPK2 and MAPKAPK3, as well as transcription factors such as ELK1 and ATF2.
The study also investigated IL-17A’s role in JNK signaling and found that IL-17A significantly elevated the downstream JNK mediators MAPK8/JNK, MAPK9/JNK2, and MAPK10/JNK3 on day 7. Additionally, MAP4 kinases RAC1 and PAK1 were upregulated in the IL-17A-treated groups, along with increased expression of MAP3 kinases and dual-specificity kinases such as MAP3K4, MAP2K7, and MAP2K4. Subsequently, transcription factors such as ELK-1, c-Jun, and ATF-2, which are involved in this pathway, were also elevated in both groups.
The study further evaluated the involvement of MAPKs in the modulation of osteogenic differentiation of SHEDs induced with IL-17A by assessing the expression of essential transcription factors in osteogenic differentiation, including COL1A1, RUNX2, and ALP. Remarkable increases were observed in the IL-17A-treated groups, indicating that MAPK signaling was activated during SHEDs’ osteogenic differentiation.
IL-17A promoted SHED osteogenic differentiation via ERK/MAPK signaling pathway
The analysis of the ERK/MAPK signaling pathway in SHEDs was conducted by culturing the cells in osteoinducing medium and treating them with IL-17A at a concentration of 50 ng/mL. The expression of ERK and p-ERK during osteogenic differentiation was evaluated and normalized using GAPDH as an endogenous protein, as shown in Figure 5.
The results showed that the expression of ERK was detected in both the SHED + OM and SHED + OM + IL-17A groups on days 1, 3, 7, 10, and 14 of osteogenic differentiation. The highest expression of ERK was observed on day 14 in the SHED + OM + IL-17A group. Additionally, the expression of p-ERK showed an increasing trend during the osteogenic differentiation phase in both the SHED + OM and SHED + OM + IL-17A groups, with the highest expression observed on day 10 in the SHED + OM + IL-17A group.
Further analysis of ERK expression revealed that it gradually increased throughout the osteogenic differentiation phase and peaked during the mineralization phase on day 14, with significant increases on days 10 and 14 in the SHED + OM + IL-17A group compared to day 1 of SHED + OM (p < 0.05). The highest expression of ERK was demonstrated in the SHED + OM + IL-17A group on day 14.
Similarly, the expression of p-ERK increased gradually from days 1 to 10 in both the SHED + OM and SHED + OM + IL-17A groups, peaking during the differentiation phase on day 10, as shown in Figure 5C. The highest expression of p-ERK was observed on day 10 in the SHED + OM + IL-17A group (p < 0.05) compared to days 1, 3, and 7 in the SHED + OM group. Remarkably, the expression of p-ERK was significantly upregulated on day 14 in the SHED + OM + IL-17A group compared to day 1 in the SHED + OM group (p < 0.05).
Discussion
SHEDs were used in this study due to their numerous advantages, which include robust stemness, pluripotency, relative ease of harvesting, and clinical importance20, 21. The osteogenic differentiation potential of SHEDs has been well-documented. On the other hand, the underlying mechanism of IL-17A modulation and its osteogenic function in SHEDs remain unknown. A deeper understanding of this mechanism will thus be essential for SHEDs’ high-performing stimulation of osteogenic differentiation. This study demonstrated the potential function of IL-17A in the osteogenic differentiation and proliferation mechanisms of SHEDs grown in two different culture conditions. Moreover, both the IL-17A and MAPK signaling pathways are essential for cell differentiation and proliferation22. Until now, no research in the literature has presented the role of the MAPK signaling pathway within the osteogenic regulation of IL-17A in SHEDs. In this study, we demonstrated that IL-17A and the MAPK signaling pathway are involved in the proliferation and differentiation of SHEDs.
In addition, Western blot analyses of OPN, COL1, OCN, ALP, and RUNX2 markers were carried out to assess the osteogenic differentiation induced by IL-17A in SHEDs. ALP is widely known as an early-stage osteogenic marker23. Type 1 collagen is one of the major components of bone extracellular matrix24. The development of inorganic bone matrix is associated with COL1, which is regarded as a prognostic marker of osteoblast differentiation and constitutes roughly 90% of its organic matrix25. RUNX2 is an essential transcription factor for MSCs to differentiate into osteogenic cells26. The osteogenic marker genes BSP, ALP, type 1 collagen, and osteocalcin can be expressed under RUNX2 regulation27. OCN, which is expressed in the last stages of osteogenic differentiation, is the top prevalent non-collagenous bone matrix protein28. It is also a prominent marker of late-stage osteoblast differentiation29. The RUNX2 expression level was elevated during the early differentiation stage, whereas OPN, COL1, OCN, and ALP expression levels were elevated in the late differentiation stages. Moreover, in SHEDs supplicated with IL-17A, all markers were expressed at elevated levels.
The correlation of both extracellular signals and transcriptional regulation is critical for controlling cell lineage selection. MAPK pathways moniker non-canonical pathways30 regulate several important signaling events intracellularly, such as cell growth, apoptosis, proliferation, and differentiation31, 32. Furthermore, MAPK’s role in the osteogenic differentiation of MSCs has been well-documented33, 34, 35. Despite becoming stimulated by diverse stimuli, the key members of this cascade, p38, ERK5, JNKs, and ERK1/2, have basically distinct downstream targets and illustrate their specific functions in various cellular functions36. Although the central role of IL-17A in the modulation of numerous biological mechanisms in somatic tissues is already established, research into the role of IL-17 in cell differentiation is in its infancy. Furthermore, IL-17A has been discovered to stimulate all three subgroups of MAPKs in a cell-specific manner across various cell types37, 38, 39. Previous research in mouse myoblasts11 and human bone marrow MSCs12 showed that MAPK and ERK1/2 were involved in the IL-17-mediated enhancement of osteogenic differentiation12. On the other hand, according to Dordevic et al.15, IL-17 significantly inhibited the osteogenic development of PDLSCs via the ERK1/2 and JNK/MAPK signaling pathways.
Several studies have reported the association between ERK1/2 and osteogenic markers. Among these, a study by Zhang et al. (2016)40 stated that the OPN and COL1A expressions increased when a low magnitude of mechanical stress (LMMS) was applied, but the expressions were significantly inhibited when treated with a MEK1/2 inhibitor. They also analyzed the involvement of LMMS-activated ERK in osteoblast differentiation; ALP activity was significantly increased by the application of LMMS40. Similar to the role of LMMS in that study, this study also demonstrated a notable effect of IL-17A on SHEDs concerning OPN and COL1A1 regulation, as well as ALP activity by stimulating the ERK pathway, which promoted osteogenesis.
According to Kocic et al., the induction of the osteogenesis process by IL-17 was correlated with an increase in the level of Runx2 mRNA11. Herein, IL-17A increased SHEDs’ osteogenic differentiation. IL-17A significantly increased RUNX2 expression and activated ERK signaling in SHEDs. These findings suggest that an ERK/RUNX2-dependent pathway mediates IL-17A-induced SHED osteogenesis. Our findings support the notion that VEGF-C would stimulate MSC osteogenesis through a RUNX2- and ERK-dependent signaling pathway31. Previous research supports our findings that Runx2 is regulated by ERK/MAPK, implying that this cascade plays a significant role in modulating the expression of osteoblast-specific genes41, 42, 43.
Recently, Wu et al. demonstrated that TRAF4-ERK5 signaling is critical for epidermal cell proliferation stimulated by IL-17A44. The ERK5 cascade is associated with the corresponding kinases included in our study: MAP3K1, MAP3K2, MAP2K5, MAPK7, and MAP3K3, which is induced by IL-17A. MAP3K2, MAP3K3, and MAP3K1 45, 46, 47 are serine/threonine protein kinases that can activate and phosphorylate MAP2K5, leading to the activation of MAPK7, belonging to the MAP kinase family. This is involved in the modulation of cell differentiation, proliferation, and survival. Mitogen-activated protein kinase 5 (MAP2K5/MEK5) activation phosphorylates various downstream targets, including MEF2C48, 49. IL-17A activated all of the ERK5 kinases in our study, which modulated the osteogenic differentiation and proliferation of SHEDs.
Gu et al. (2015) evaluated the function of c-Jun N-terminal kinase (c-JNK) in the osteogenic differentiation of human adipose-derived MSCs50. The suppression of the JNK signaling cascade inhibited osteogenic differentiation in a dose-dependent manner, as demonstrated by combining an extracellular calcium deposition identification, ALP activity assay, and osteogenesis-associated genes (ALP, OCN, and Runx2). The authors concluded that the JNK pathway regulated early and late osteogenic differentiation of hAMSCs by modulating ALP, RUNX2, and OCN expression and matrix mineralization. These findings are similar to our study of SHEDs, wherein the corresponding protein and gene expressions were enhanced by IL-17A through JNK activation. Based on these observations, we propose that the c-JNK pathway promotes osteogenesis in SHEDs.
These findings point to a potential mechanism in which the increased expression of all MAPK, MKKK, and MKK family members, genes in response to IL-17A stimulation, and transcription factors are modulated by the MAPK cascade. This provides a novel insight into the molecular mechanisms of IL-17A-regulated SHEDs. IL-17A has received considerable attention in the pathogenesis of periodontitis, primarily due to its ability to promote inflammation and enhance protective antimicrobial immune responses51. Consequently, targeting IL-17 or its receptor has emerged as a potential strategy to modulate MAPK activation and alleviate the inflammatory response associated with periodontitis.
In our study, the osteogenesis effect of IL-17 was apparently dependent on p38, ERK1/2, and JNK MAPK activity. Notably, expression of all ERK, p38, and JNK upstream and downstream signals was likewise elevated, as was the increase of both osteogenic genes and COL1A1 included in this array. The increase in COL1A1 levels highlighted that IL-17A can induce SHED osteogenic differentiation via MAPK activation.
The ERK expression level of SHED + OM + IL-17A on day 10 significantly increased and showed the highest peak on day 14 when compared to day 1 of SHED + OM. This result demonstrated that ERK expression was only upregulated during several stages of SHED osteogenic differentiation, such as differentiation (day 10) and the mineralization phase (day 14). In addition, these findings suggested that IL-17A played a significant role in enhancing the osteogenic differentiation process of SHED, corresponding with the ERK expression level in the IL-17A-treated group. This was supported by a study stating that insoluble and soluble extracellular signals could stimulate ERK1/2 and lead to the activation of crucial cellular processes such as essential transcriptional and phenotypic differentiating pathways via the protein kinase cascade activation43.
In this study, p-ERK expression was evaluated in SHED, SHED + OM, and SHED + OM + IL-17A, specifically recognizing the dually phosphorylated (fully activated) ERK signaling pathway in the SHED osteogenic differentiation process. The p-ERK expression showed markedly increased expression from day 1 until day 10 and decreased during the mineralization phase (day 14) in SHED + OM and SHED + OM + IL-17A. Our result demonstrated significant upregulation, with the highest peak of p-ERK expression during the differentiation phase at day 10 in SHED + OM + IL-17A when compared to day 1 in SHED + OM. This was supported by a recent study that showed that ERK phosphorylation levels peaked at osteogenic culture day 12 and slowly decreased in late differentiation phases, indicating that ERK was highly elevated in mature osteoblasts51. However, no significant p-ERK expression occurred in either SHED + OM group on days 1, 3, or 7, which might be due to the early osteogenic differentiation phase in SHEDs. A previous study stated that the bone regulation mechanism was usually undetectable on day 352. Interestingly, that study confirmed that undifferentiated SHEDs did not activate the ERK signaling pathway. The activation of ERK1/2 signaling pathways in the osteogenic differentiation of SHEDs supplicated with IL-17A was similar to the previous study that showed that IL-17A enhanced osteogenic differentiation in IL-17A-supplicated SHEDs in comparison to untreated cells by demonstrating increased ALP, OPN, RUNX2, OCN, and COL1A1 expression. These findings were also supported by another study that stated that the ERK/MAPK pathway indirectly affected osteoblast differentiation by activating RSK2, which subsequently phosphorylated the transcriptional regulator of late-stage osteoblast synthetic functions, ATF453. Further studies on the effect of IL-17A in in vivo experiments will further elucidate the involvement of cytokines in bone regulation.
Conclusions
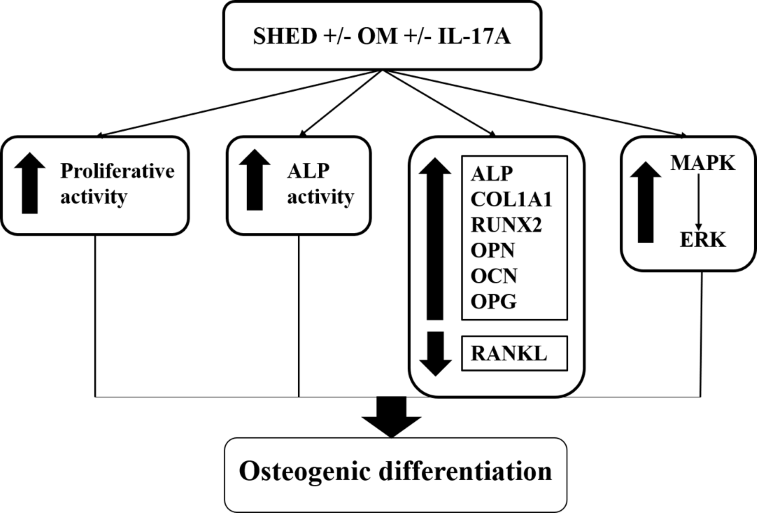
These findings highlight the significance of the ERK/MAPK signaling cascade in mediating the osteogenic differentiation of SHEDs in response to IL-17A stimulation (Figure 6). These results emphasize the important role of these intracellular signaling pathways in regulating the osteogenic potential of SHEDs. This has significant implications for the potential application of SHEDs in bone tissue engineering, especially considering the wide range of cellular sources that can be used for this purpose.
Abbreviations
BSP: Bone sialoprotein, Colla1: Type I collagen, IL-17A: Interleukin-17A, MAPK: Mitogen-activated protein kinases, NF-κB: Nuclear factor-κB, OPG: Osteoprotegerin, OPN: Osteopontin, SHED: Stem cells isolated from human exfoliated deciduous teeth, Th17: T helper 17 cell, α-MEM: Alpha minimum essential medium
Acknowledgments
The authors thank the staff of the Craniofacial Sciences Laboratory, School of Dental Sciences and the Advanced Molecular Laboratory, School of Health Sciences Universiti Sains Malaysia for their help rendered in this study.
Author’s contributions
Conceptualization, AAN; Formal analysis, AAS and AAN; Funding acquisition, AAN; Investigation, MNRAM, AAS and AAN; Methodology, AAS and AAN; Supervision, KTP and AAN; Validation, AAN; Writing – original draft, MNRAM and AAS; Writing – review & editing, KTP, NHS and AAN. All authors read and approved the final manuscript.
Funding
This research was funded by the Fundamental Research Grant Scheme from the Ministry of Higher Education, Malaysia (FRGS/1/2018/SKK08/USM/02/12).
Availability of data and materials
The dataset used and/or analysed in the current study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Ansari
M.,
Bone tissue regeneration: biology, strategies and interface studies. Progress in Biomaterials.
2019;
8
(4)
:
223-37
.
View Article PubMed Google Scholar -
Zhang
T.,
Yao
Y.,
Effects of inflammatory cytokines on bone/cartilage repair. Journal of Cellular Biochemistry.
2019;
120
(5)
:
6841-50
.
View Article PubMed Google Scholar -
Korn
T.,
Bettelli
E.,
Oukka
M.,
Kuchroo
V.K.,
IL-17 and Th17 Cells. Annual Review of Immunology.
2009;
27
(1)
:
485-517
.
View Article PubMed Google Scholar -
Toy
D.,
Kugler
D.,
Wolfson
M.,
Vanden Bos
T.,
Gurgel
J.,
Derry
J.,
Cutting edge: interleukin 17 signals through a heteromeric receptor complex. Journal of Immunology (Baltimore, Md.: 1950).
2006;
177
(1)
:
36-9
.
View Article PubMed Google Scholar -
Shen
F.,
Gaffen
S.L.,
Structure-function relationships in the IL-17 receptor: implications for signal transduction and therapy. Cytokine.
2008;
41
(2)
:
92-104
.
View Article PubMed Google Scholar -
Patel
D.N.,
King
C.A.,
Bailey
S.R.,
Holt
J.W.,
Venkatachalam
K.,
Agrawal
A.,
Interleukin-17 stimulates C-reactive protein expression in hepatocytes and smooth muscle cells via p38 MAPK and ERK1/2-dependent NF-kappaB and C/EBPbeta activation. The Journal of Biological Chemistry.
2007;
282
(37)
:
27229-38
.
View Article PubMed Google Scholar -
Li
X.,
Bechara
R.,
Zhao
J.,
McGeachy
M.J.,
Gaffen
S.L.,
IL-17 receptor–based signaling and implications for disease. Nature immunology.
2019;
20
(12)
:
1594-1602
.
View Article Google Scholar -
Vernal
R.,
Dutzan
N.,
Chaparro
A.,
Puente
J.,
Antonieta Valenzuela
M.,
Gamonal
J.,
Levels of interleukin-17 in gingival crevicular fluid and in supernatants of cellular cultures of gingival tissue from patients with chronic periodontitis. Journal of Clinical Periodontology.
2005;
32
(4)
:
383-9
.
View Article PubMed Google Scholar -
Benham
H.,
Norris
P.,
Goodall
J.,
Wechalekar
M.D.,
FitzGerald
O.,
Szentpetery
A.,
Th17 and Th22 cells in psoriatic arthritis and psoriasis. Arthritis Research & Therapy.
2013;
15
(5)
:
136
.
View Article PubMed Google Scholar -
Jansen
D.T.,
Hameetman
M.,
van Bergen
J.,
Huizinga
T.W.,
van der Heijde
D.,
Toes
R.E.,
IL-17-producing CD4+ T cells are increased in early, active axial spondyloarthritis including patients without imaging abnormalities. Rheumatology (Oxford, England).
2015;
54
(4)
:
728-35
.
View Article PubMed Google Scholar -
Kocić
J.,
Santibañez
J.F.,
Krstić
A.,
Mojsilović
S.,
Dor\djević
I.O.,
Trivanović
D.,
Interleukin 17 inhibits myogenic and promotes osteogenic differentiation of C2C12 myoblasts by activating ERK1,2. Biochimica et Biophysica Acta.
2012;
1823
(4)
:
838-49
.
View Article PubMed Google Scholar -
Osta
B.,
Lavocat
F.,
Eljaafari
A.,
Miossec
P.,
Effects of interleukin-17A on osteogenic differentiation of isolated human mesenchymal stem cells. Frontiers in Immunology.
2014;
5
:
425
.
View Article PubMed Google Scholar -
Jo
S.,
Wang
S.E.,
Lee
Y.L.,
Kang
S.,
Lee
B.,
Han
J.,
IL-17A induces osteoblast differentiation by activating JAK2/STAT3 in ankylosing spondylitis. Arthritis Research & Therapy.
2018;
20
(1)
:
115
.
View Article PubMed Google Scholar -
Wang
Z.,
Jia
Y.,
Du
F.,
Chen
M.,
Dong
X.,
Chen
Y.,
IL-17A inhibits osteogenic differentiation of bone mesenchymal stem cells via Wnt signaling pathway. Medical Science Monitor.
2017;
23
:
4095-101
.
View Article PubMed Google Scholar -
\DJor\djević
I.O.,
Kukolj
T.,
Krstić
J.,
Trivanović
D.,
Obradović
H.,
Santibañez
J.F.,
The inhibition of periodontal ligament stem cells osteogenic differentiation by IL-17 is mediated via MAPKs. The International Journal of Biochemistry & Cell Biology.
2016;
71
:
92-101
.
View Article PubMed Google Scholar -
Liao
C.,
Zhang
C.,
Jin
L.,
Yang
Y.,
IL-17 alters the mesenchymal stem cell niche towards osteogenesis in cooperation with osteocytes. Journal of Cellular Physiology.
2020;
235
(5)
:
4466-80
.
View Article PubMed Google Scholar -
Noh
M.,
Interleukin-17A increases leptin production in human bone marrow mesenchymal stem cells. Biochemical Pharmacology.
2012;
83
(5)
:
661-70
.
View Article PubMed Google Scholar -
Su
W.T.,
Chen
X.W.,
Stem cells from human exfoliated deciduous teeth differentiate into functional hepatocyte-like cells by herbal medicine. Bio-Medical Materials and Engineering.
2014;
24
(6)
:
2243-7
.
View Article PubMed Google Scholar -
Yamaza
T.,
Kentaro
A.,
Chen
C.,
Liu
Y.,
Shi
Y.,
Gronthos
S.,
Immunomodulatory properties of stem cells from human exfoliated deciduous teeth. Stem Cell Research & Therapy.
2010;
1
(1)
:
5
.
View Article PubMed Google Scholar -
Miura
M.,
Gronthos
S.,
Zhao
M.,
Lu
B.,
Fisher
L.W.,
Robey
P.G.,
SHED: stem cells from human exfoliated deciduous teeth. Proceedings of the National Academy of Sciences of the United States of America.
2003;
100
(10)
:
5807-12
.
View Article PubMed Google Scholar -
Rosa
V.,
Dubey
N.,
Islam
I.,
Min
K.S.,
Nör
J.E.,
Pluripotency of stem cells from human exfoliated deciduous teeth for tissue engineering. Stem Cells International.
2016;
2016
:
5957806
.
View Article PubMed Google Scholar -
Jiang
L.,
Tang
Z.,
Expression and regulation of the ERK1/2 and p38 MAPK signaling pathways in periodontal tissue remodeling of orthodontic tooth movement. Molecular Medicine Reports.
2018;
17
(1)
:
1499-506
.
PubMed Google Scholar -
Sun
L.,
Wu
L.,
Bao
C.,
Fu
C.,
Wang
X.,
Yao
J.,
Gene expressions of Collagen type I, ALP and BMP-4 in osteo-inductive BCP implants show similar pattern to that of natural healing bones. Materials Science and Engineering C.
2009;
29
(6)
:
1829-34
.
View Article Google Scholar -
Gelse
K.,
Pöschl
E.,
Aigner
T.,
Collagens-structure, function, and biosynthesis. Advanced Drug Delivery Reviews.
2003;
55
(12)
:
1531-46
.
View Article PubMed Google Scholar -
Osteoporosis in men: the effects of gender on skeletal healthAcademic press 2009.
Google Scholar -
Artigas
N.,
Ureña
C.,
Rodríguez-Carballo
E.,
Rosa
J.L.,
Ventura
F.,
Mitogen-activated protein kinase (MAPK)-regulated interactions between Osterix and Runx2 are critical for the transcriptional osteogenic program. The Journal of Biological Chemistry.
2014;
289
(39)
:
27105-17
.
View Article PubMed Google Scholar -
Liu
T.M.,
Lee
E.H.,
Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Engineering. Part B, Reviews.
2013;
19
(3)
:
254-63
.
View Article PubMed Google Scholar -
Chen
Q.,
Liu
W.,
Sinha
K.M.,
Yasuda
H.,
de Crombrugghe
B.,
Identification and characterization of microRNAs controlled by the osteoblast-specific transcription factor Osterix. PLoS One.
2013;
8
(3)
:
e58104
.
View Article PubMed Google Scholar -
McKee
M.D.,
Addison
W.N.,
Kaartinen
M.T.,
Hierarchies of extracellular matrix and mineral organization in bone of the craniofacial complex and skeleton. Cells, Tissues, Organs.
2005;
181
(3-4)
:
176-88
.
View Article PubMed Google Scholar -
Xu
D.J.,
Zhao
Y.Z.,
Wang
J.,
He
J.W.,
Weng
Y.G.,
Luo
J.Y.,
Smads, p38 and ERK1/2 are involved in BMP9-induced osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. BMB Reports.
2012;
45
(4)
:
247-52
.
View Article PubMed Google Scholar -
Chan
Y.H.,
Ho
K.N.,
Lee
Y.C.,
Chou
M.J.,
Lew
W.Z.,
Huang
H.M.,
Melatonin enhances osteogenic differentiation of dental pulp mesenchymal stem cells by regulating MAPK pathways and promotes the efficiency of bone regeneration in calvarial bone defects. Stem Cell Research & Therapy.
2022;
13
(1)
:
73
.
View Article PubMed Google Scholar -
Mu
R.,
Chen
B.,
Bi
B.,
Yu
H.,
Liu
J.,
Li
J.,
Lim mineralization protein-1 enhances the committed differentiation of dental pulp stem cells through the erk1/2 and p38 mapk pathways and bmp signaling. International Journal of Medical Sciences.
2022;
19
(8)
:
1307-19
.
View Article PubMed Google Scholar -
Li
C.S.,
Zheng
Z.,
Su
X.X.,
Wang
F.,
Ling
M.,
Zou
M.,
Activation of the extracellular signal-regulated kinase signaling is critical for human umbilical cord mesenchymal stem cell osteogenic differentiation. BioMed Research International.
2016;
2016
:
3764372
.
View Article PubMed Google Scholar -
Xu
C.,
Zheng
Z.,
Fang
L.,
Zhao
N.,
Lin
Z.,
Liang
T.,
Phosphatidylserine enhances osteogenic differentiation in human mesenchymal stem cells via ERK signal pathways. Materials Science and Engineering C.
2013;
33
(3)
:
1783-8
.
View Article PubMed Google Scholar -
Murakami
J.,
Ishii
M.,
Suehiro
F.,
Ishihata
K.,
Nakamura
N.,
Nishimura
M.,
Vascular endothelial growth factor-C induces osteogenic differentiation of human mesenchymal stem cells through the ERK and RUNX2 pathway. Biochemical and Biophysical Research Communications.
2017;
484
(3)
:
710-8
.
View Article PubMed Google Scholar -
Gaffen
S.L.,
An overview of IL-17 function and signaling. Cytokine.
2008;
43
(3)
:
402-7
.
View Article PubMed Google Scholar -
Krstić
A.,
Ilić
V.,
Mojsilović
S.,
Jovcić
G.,
Milenković
P.,
Bugarski
D.,
p38 MAPK signaling mediates IL-17-induced nitric oxide synthase expression in bone marrow cells. Growth Factors (Chur, Switzerland).
2009;
27
(2)
:
79-90
.
View Article PubMed Google Scholar -
Ivanov
S.,
Lindén
A.,
Interleukin-17 as a drug target in human disease. Trends in Pharmacological Sciences.
2009;
30
(2)
:
95-103
.
View Article PubMed Google Scholar -
Xu
S.,
Cao
X.,
Interleukin-17 and its expanding biological functions. Cellular & Molecular Immunology.
2010;
7
(3)
:
164-74
.
View Article PubMed Google Scholar -
Zhang
Q.,
Matsui
H.,
Horiuchi
H.,
Liang
X.,
Sasaki
K.,
A-Raf and C-Raf differentially regulate mechanobiological response of osteoblasts to guide mechanical stress-induced differentiation. Biochemical and Biophysical Research Communications.
2016;
476
(4)
:
438-44
.
View Article PubMed Google Scholar -
Ge
C.,
Xiao
G.,
Jiang
D.,
Yang
Q.,
Hatch
N.E.,
Roca
H.,
Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. The Journal of Biological Chemistry.
2009;
284
(47)
:
32533-43
.
View Article PubMed Google Scholar -
Deschaseaux
F.,
Sensébé
L.,
Heymann
D.,
Mechanisms of bone repair and regeneration. Trends in Molecular Medicine.
2009;
15
(9)
:
417-29
.
View Article PubMed Google Scholar -
Liu
Q.,
Cen
L.,
Zhou
H.,
Yin
S.,
Liu
G.,
Liu
W.,
The role of the extracellular signal-related kinase signaling pathway in osteogenic differentiation of human adipose-derived stem cells and in adipogenic transition initiated by dexamethasone. Tissue Engineering. Part A.
2009;
15
(11)
:
3487-97
.
View Article PubMed Google Scholar -
Wu
L.,
Chen
X.,
Zhao
J.,
Martin
B.,
Zepp
J.A.,
Ko
J.S.,
A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. The Journal of Experimental Medicine.
2015;
212
(10)
:
1571-87
.
View Article PubMed Google Scholar -
Chao
T.H.,
Hayashi
M.,
Tapping
R.I.,
Kato
Y.,
Lee
J.D.,
MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. The Journal of Biological Chemistry.
1999;
274
(51)
:
36035-8
.
View Article PubMed Google Scholar -
Sun
W.,
Kesavan
K.,
Schaefer
B.C.,
Garrington
T.P.,
Ware
M.,
Johnson
N.L.,
MEKK2 associates with the adapter protein Lad/RIBP and regulates the MEK5-BMK1/ERK5 pathway. The Journal of Biological Chemistry.
2001;
276
(7)
:
5093-100
.
View Article PubMed Google Scholar -
Xu
B.E.,
Stippec
S.,
Lenertz
L.,
Lee
B.H.,
Zhang
W.,
Lee
Y.K.,
WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. The Journal of Biological Chemistry.
2004;
279
(9)
:
7826-31
.
View Article PubMed Google Scholar -
Nishimoto
S.,
Nishida
E.,
MAPK signalling: ERK5 versus ERK1/2. EMBO Reports.
2006;
7
(8)
:
782-6
.
View Article PubMed Google Scholar -
Roberts
O.L.,
Holmes
K.,
Müller
J.,
Cross
D.A.,
Cross
M.J.,
ERK5 and the regulation of endothelial cell function. Biochemical Society Transactions.
2009;
37
(Pt 6)
:
1254-9
.
View Article PubMed Google Scholar -
Gu
H.,
Huang
Z.,
Yin
X.,
Zhang
J.,
Gong
L.,
Chen
J.,
Role of c-Jun N-terminal kinase in the osteogenic and adipogenic differentiation of human adipose-derived mesenchymal stem cells. Experimental Cell Research.
2015;
339
(1)
:
112-21
.
View Article PubMed Google Scholar -
Kim
J.M.,
Yang
Y.S.,
Park
K.H.,
Oh
H.,
Greenblatt
M.B.,
Shim
J.H.,
The ERK MAPK pathway is essential for skeletal development and homeostasis. International Journal of Molecular Sciences.
2019;
20
(8)
:
1803
.
View Article PubMed Google Scholar -
Shim
J.H.,
Greenblatt
M.B.,
Zou
W.,
Huang
Z.,
Wein
M.N.,
Brady
N.,
Schnurri-3 regulates ERK downstream of WNT signaling in osteoblasts. The Journal of Clinical Investigation.
2013;
123
(9)
:
4010-22
.
View Article PubMed Google Scholar -
Ai-Aql
Z.S.,
Alagl
A.S.,
Graves
D.T.,
Gerstenfeld
L.C.,
Einhorn
T.A.,
Molecular mechanisms controlling bone formation during fracture healing and distraction osteogenesis. Journal of Dental Research.
2008;
87
(2)
:
107-18
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 10 No 5 (2023)
Page No.: 5686-5700
Published on: 2023-05-31
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 3685 times
- PDF downloaded - 1496 times
- XML downloaded - 158 times
 Biomedpress
Biomedpress








