Abstract
Introduction: Existing research establishes the role of oligodeoxynucleotides (ODNs) containing cytosine-phosphate-guanine motifs (CpG ODNs) as adjuvants to vaccines against infectious diseases. However, the natural structure of ODN with phosphodiester (PD) linkages is prone to degradation by serum DNase, which limits the application of CpG ODNs. We recently engineered a monomeric guanine-quadruplex (G4)-structured CpG ODN (G4 CpG ODN) and determined that the monomeric G4 increased the DNase resistance, intracellular uptake, and cytokine induction of CpG ODN in vitro and in vivo. However, the effect of the primary sequence on G4 conformation and function of G4 CpG ODN is unclear.
Methods: Therefore, in this study, we examine the role of loop permutation, a sequential order of three G4 loops, in G4 folding and immunostimulatory properties of G4 CpG ODNs. We explore the impact of loop permutation on the immunostimulatory activity of GD3_2L, a monomeric G4 CpG ODN that we reported previously. We exchanged the sequence of the second loop with the first and third loops of GD3_2L to obtain GD3_1L and GD3_3L, respectively. The structure of the obtained ODNs was determined by circular dichroism and polyacrylamide gel electrophoresis; the thermodynamic stability of ODN was evaluated through UV melting curve analysis. We assessed the immunostimulatory activity by examining the relative messenger RNA (mRNA) levels of cytokines produced in G4 CpG ODN-stimulated mouse macrophage-like RAW264 by reverse transcription-quantitative polymerase chain reaction method.
Results: We found that changing the sequential order of CpG motifs from the second to the first loop resulted in a change in G4 conformation from hybrid to parallel and a decrease in the compactness and thermodynamic stability of GD3_2L. Moreover, swapping order between the second and third loop of GD3_2L led to a hybrid G4 structure that is as stable as GD3_2L but possesses a lower cytokine induction.
Conclusion: In conclusion, the loop permutation does not always change the topology and structural compactness; instead, it changes the conformation of CpG motifs in the G4 structure, directly affecting the immunostimulatory properties. Our result demonstrates that the loop-transposition-mediated structural compactness decrease can, if exploited, increase the efficacy of immunostimulatory G4 CpG ODN.
Introduction
Recently developed vaccines are not based on the entire organism but on its antigenic (Ag) parts, such as protein, DNA, or RNA. These vaccines show poor immunogenicity and, thus, difficulty in inducing effective immune responses. These vaccines also require including a vaccine adjuvant to trigger the innate immune response and allow the vaccine to induce an Ag-specific immunity at a high level and for a long duration1. Oligodeoxynucleotides (ODNs) containing cytosine-phosphate-guanine (CpG) motifs represent new-generation vaccine adjuvants. CpG motifs such as GTCGTT or GACGTT are abundant in bacterial and viral DNA but are found in very low frequencies or methylated in vertebrate DNA. CpG ODN is thus a potent stimulator of the innate immune system through the activation of toll-like receptor 9 (TLR9), which is one of the pattern recognition receptors of the innate immune system2. However, native ODN with a phosphodiester (PD) backbone is susceptible to degradation by serum DNase, which interferes with therapeutic applications. Consequently, phosphorothioate backbone-based CpG ODNs (PT CpG ODNs) have been investigated as TLR9 agonists. CpG 1018, a PT CpG ODN, has been used as an adjuvant in the hepatitis B vaccine HEPLISAV-B recently approved by the United States Food and Drug Administration (US FDA). However, PT ODNs can reportedly cause thrombocytopenia, anemia, and leukopenia. Furthermore, there is still concern about the long-term side effects of PT ODNs, such as neurodegenerative diseases (e.g., Alzheimer’s disease), because the PT backbone has a high nonspecific affinity toward various proteins3. Hence, improving the DNase stability of CpG ODNs without chemically denaturing their backbone has attracted significant attention from researchers.
We have reported that using a monomeric guanine-quadruplex (G4) structure as a scaffold can increase DNase resistance and immunostimulatory efficacy of linear PD CpG ODNs (Figure 1)4. The G4 structure is a non-canonical DNA structure that is highly stable and exists naturally in guanine-rich regions of DNA, such as in the telomeres of eukaryotic chromosomes and the promoter region. The monomeric G4 CpG ODN consists of a G4 stem and three single-stranded loops. The G4 stem is composed of stacked guanine quartets, where the guanine quartet comprises four guanines arranged in a square plane and stabilized by Hoogsteen hydrogen bonds (Figure 1). Single-stranded loops connect the guanine quartets in the G4 stem. The CpG motifs in the single-stranded loops are capable of activating TLR9 and providing immunostimulatory activities4, 5.
We reported that GD3_2L, a G4 CpG ODN containing three CpG motifs in the second loop, induced more robust production of interleukin (IL)-6, IL-12, and interferon (IFN)-β in mouse macrophage-like RAW264 cells than GD1, and GD2, which are the G4 CpG ODNs containing one and two CpG motifs, respectively4. In an attempt to further increase the immunostimulatory properties of GD3_2L, we explored the impact of specific factors on the efficacy of GD3_2L. We found that the combination with cationic lipid6 or G4-binding ligand7 allows GD3_2L to induce higher cytokine production in mouse macrophage-like RAW264 cells than naïve GD3_2L. However, the influence of loop mutation on GD3_2L efficacy remains to be clarified. In addition, the position of the long loop can affect the folding of G4 ODNs, which may influence the function of G4 ODNs8. Therefore, in this study, we analyzed the effects of the position of the loop containing CpG motifs on the immunostimulatory activity of GD3_2L while maintaining identical loop compositions. The use of circular dichroism (CD) and polyacrylamide gel electrophoresis (PAGE) determined the ODN’s structure and implementing UV melting curve analysis determined the ODN’s thermodynamic stability. We assessed the immunostimulatory activity by examining the relative messenger RNA (mRNA) levels of cytokines produced in G4 CpG ODN-stimulated mouse macrophage-like RAW264 by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
| Name | Sequence (from 5’ to 3’) |
|---|---|
| GD3_1L | GGG G T C G T T ¯ TT G T C G T T ¯ TT G T C G T T ¯ GGG TT GGG TT GGG |
| GD3_2L | GGG TT GGG G T C G T T ¯ TT G T C G T T ¯ TT G T C G T T ¯ GGG TT GGG |
| GD3_3L | GGG TT GGG TT GGG G T C G T T ¯ TT G T C G T T ¯ TT G T C G T T ¯ GGG |
Methods
Oligodeoxynucleotides
This study’s purchase source for all ODNs is Eurofins Genomics (Tokyo, Japan). The ODNs used have phosphodiester backbones and are HPLC-purified grade. Table 1 displays the ODN sequences.
G-quadruplex structure preparation of ODNs
The stock solutions (~100 μM) were prepared by dissolving the lyophilized powder of the purchased ODNs in sterile Milli-Q water and stored at −30 °C until use. Before use, the ODNs were diluted from the stock solution using Dulbecco’s phosphate-buffered saline (D-PBS, DS Pharma Biomedical, Osaka, Japan). Next, they underwent a heat treatment using a thermal cycler (PCR Thermal Cycler Dice Standard TP650, Takara Bio, Shiga, Japan) to prepare the G4 structure. For example, 2 μL of 10x D-PBS, 2 μL of 100 μM ODNs stock solution, and 16 μL of sterile Milli-Q water were mixed in a PCR tube to prepare 20 μL of 10 μM G4 CpG ODN solution. Then, the ODN solutions were heated to 95 °C for 5 min, followed by gradual cooling to 30 °C for 30 min, and then the solutions were cooled to 4 °C. Finally, we stored the G4 ODN solutions at 4 °C until use.
Circular dichroism (CD) spectroscopy
The G4 CpG ODN solutions were prepared at 20 μM in 1x D-PBS and then diluted to 2 μM with 1x D-PBS. A quartz cuvette (model T-18-ES-10, 1 cm path length, Jasco) containing 100 μL of the 2 μM ODN solution was used to measure the CD spectra. We utilized a Jasco J-725 CD spectropolarimeter to conduct the CD spectra measurements over a wavelength range of 230 to 320 nm (scan speed, 50 nm/min; response time, 2.0 s; bandwidth, 1.0 nm; resolution, 0.2 nm; and sensitivity, 100 mdeg) at 25 °C. The spectrum of each ODN is the average of five scans.
Polyacrylamide gel electrophoresis (PAGE)
Gel electrophoresis was conducted using 10 — 20% polyacrylamide gels (PAGE; 1 mm thick; ATTO, Tokyo, Japan) in tris-glycine (TG; 25 mM tris, 125 mM glycine) buffer supplemented with 4 mM KCl at a constant current of 20 mA at 4 °C. The size maker was the ultra-low range DNA ladder (Thermo Fisher Scientific, Waltham, MA, USA). In addition, SYBR gold nucleic acid gel stain (Thermo Fisher Scientific) was used to stain the bands in the gel.
CD and ultraviolet (UV) melting curves
The G4 CpG ODN solutions were prepared at 40 μM and then drawn into a quartz capillary cell (CAP-100Q, Jasco, Tokyo, Japan) via capillary action. Both ends of the capillary cell were sealed to prevent sample volatilization. Then, the cell was placed into the capillary jacket and placed into a Peltier thermostatted cell holder. A temperature sensor was inserted into the sensor insertion port on the jacket to ensure accurate sample temperature measurements. The CD and UV melting curves were determined by recording the CD and absorbance of the samples, respectively, at 260 nm and 295 nm over a temperature range of 10 to 90 °C using a Jasco J-1500 CD spectropolarimeter (Tokyo, Japan). The renaturation and denaturation and the profiles of ODNs were obtained during the cooling and reheating processes. The melting temperature (Tm) of G4 CpG ODNs was calculated from the melting curve using a previously reported method4.
Cell culture
The mouse macrophage-like RAW264 cell line (RCB0535), purchased from RIKEN BioResource Center (Tsukuba, Japan), was maintained at 37 °C in a 5% CO2-humidified incubator and grown in a minimum essential medium (MEM) (Thermo Fisher Scientific) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich, St Louis, MO, USA) and 1% (v/v) MEM nonessential amino acid solution (100x, Fujifilm Wako Pure Chemical, Osaka, Japan).
Immunostimulation of RAW264 cells
RAW264 cells in a culture medium containing 1% (v/v) penicillin/streptomycin (P/S, 100x, Thermo Fisher Scientific) were seeded into a 96-well plate (1 × 105 cells/well, 5.3 × 105 cells/mL, 190 μL) and incubated for 18 h. Then, the cells were treated with 10 μL of 80 μM ODN (the final ODN concentration is 4 μM). Afterward, the cells were incubated for 24 h before evaluating the relative transcript levels of cytokines. In addition, relative transcript levels of IL-6 and IL-12p40 in RAW264 cells were examined by RT-qPCR, as described previously4. The sequences of the forward and reverse primer of murine cytokines are as follows:
GAPDH: Forward: 5'-GTGGACCTCATGGCCTACAT-3', reverse: 5'-TGTGAGGGAGATGCTCAGTG-3'
IL6: Forward: 5'-TCCTTCCTACCCCAATTTCC-3', reverse: 5'-CGACACTAGGTTTGCCAGTA-3'
IL12p40: Forward: Forward: 5'-GAAAGGCTGGGGTATCGG-3', reverse: 5'-GGCTGTCCTCAAACTCAC-3'
Statistical analysis
Statistical significance of the differences was determined by one-way analysis of variance (ANOVA) followed by either Tukey’s multiple comparisons test for comparison with other groups or by Dunnett’s multiple comparisons test for comparison with a control group. GraphPad Prism version 9.4.0 for Windows (GraphPad Software, La Jolla, CA, USA) was used to perform all statistical analyses.
| ODNs | Melting temperature (°C) |
|---|---|
| GD3_1L | 37 |
| GD3_2L | 47 |
| GD3_3L | 44 |
Results
We designed G4 CpG ODNs containing CpG motifs at the second loop in a previous study. In the present study, to examine the influence of the position of the loop containing CpG motifs, we created GD3_1L and GD3_3L by swapping the place of the long loop containing the CpG motifs (L*) with the first (L#) and the third (L$) short TT loop of GD3_2L, respectively. These variants only differed in the order of the loop containing the CpG motif, while the length and composition were kept identical. Figure 2 shows the schematic illustration of the loop permutation of GD3_2L.
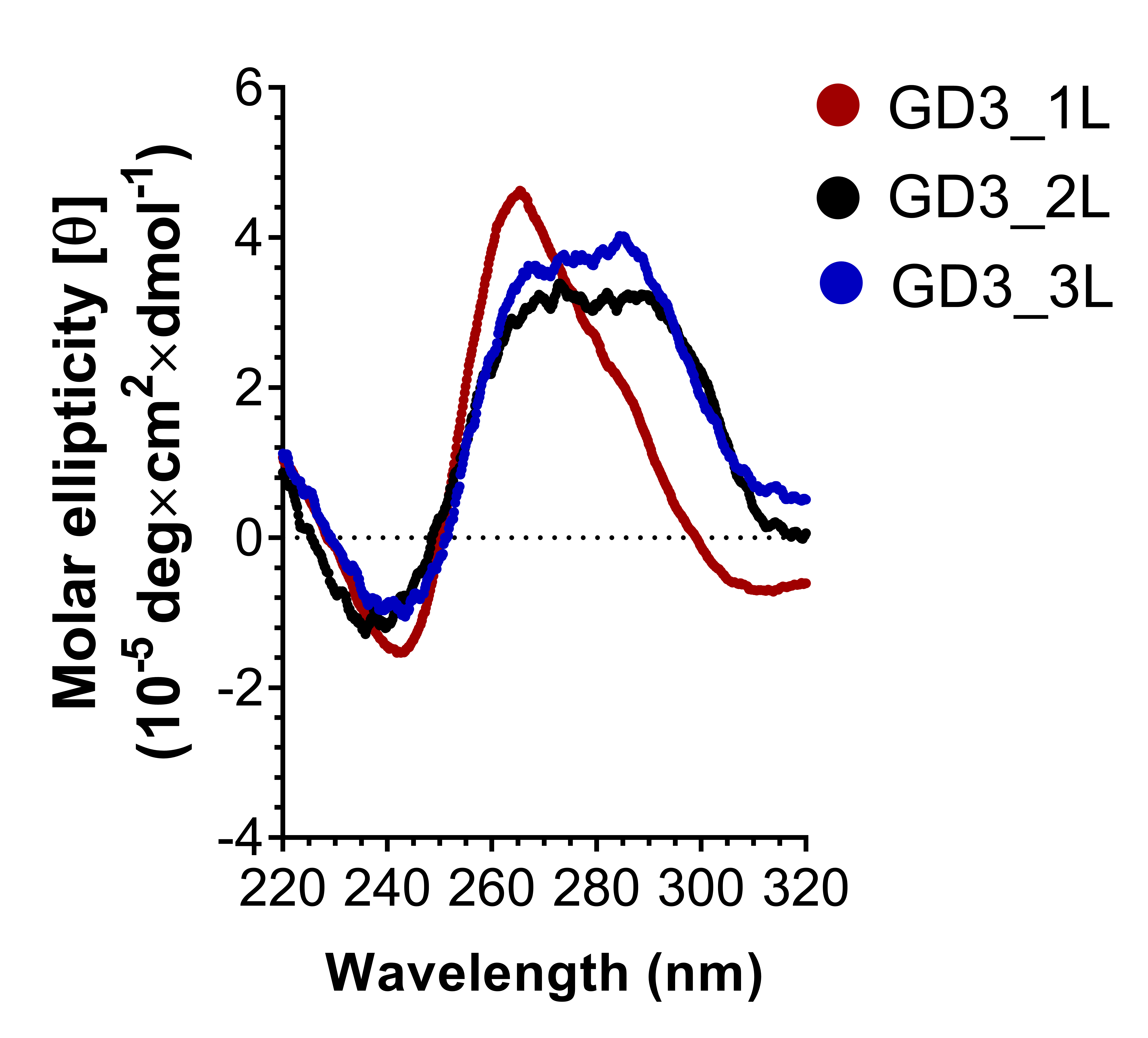
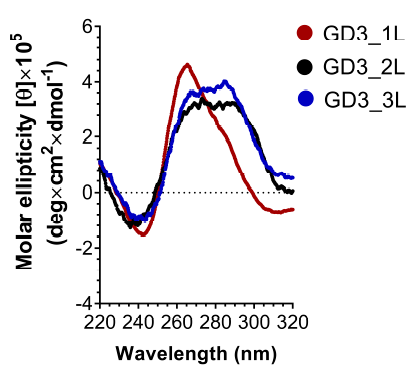
CD spectra examined the G4 topology of ODNs. Figure 3 shows that GD3_2L exhibits a negative peak at 240 nm and a broad positive peak in the 260–290 nm range, similar to the characteristic peaks of a hybrid type G4. In addition, the peak fitting and deconvolution of GD3_2L CD spectra in a previous study4 suggested that an overlap of a positive peak at around 280 nm, corresponding to the long central loop with a random structure, with two positive peaks at 260 and 295 nm, corresponding to the hybrid G4 stem, caused the broad positive peak at 260–290 nm. In addition, principal component analysis (PCA) indicated that GD3_2L forms hybrid G47. The CD spectrum of GD3_1L has a negative peak, positive peak, and bulge at 243, 266, and 285 nm, respectively, suggesting that GD3_1L mainly forms a parallel structure and partially forms a hybrid structure. GD3_3L shows a similar CD spectrum to GD3_2L, but the intensity of the broad peak of GD3_3L is slightly higher than that of GD3_2L, indicating that GD3_3L formed a hybrid G4 structure analogous to GD3_2L.
We conducted PAGE analysis of CpG ODNs in the presence of K+ in the running buffer to evaluate G4 molecularity, which is the quantity of ODN molecules forming G4. Figure 4 shows that all ODNs moved as fast as the 35-base pair (bp) marker, indicating that the ODNs are formed compact structure. GD3_1L has lower mobility than GD3_2L and GD3_3L, whereas GD3_2L and GD3_3L have similar mobility, which suggests that the GD3_1L structure is less compact than GD3_2L and GD3_3L.
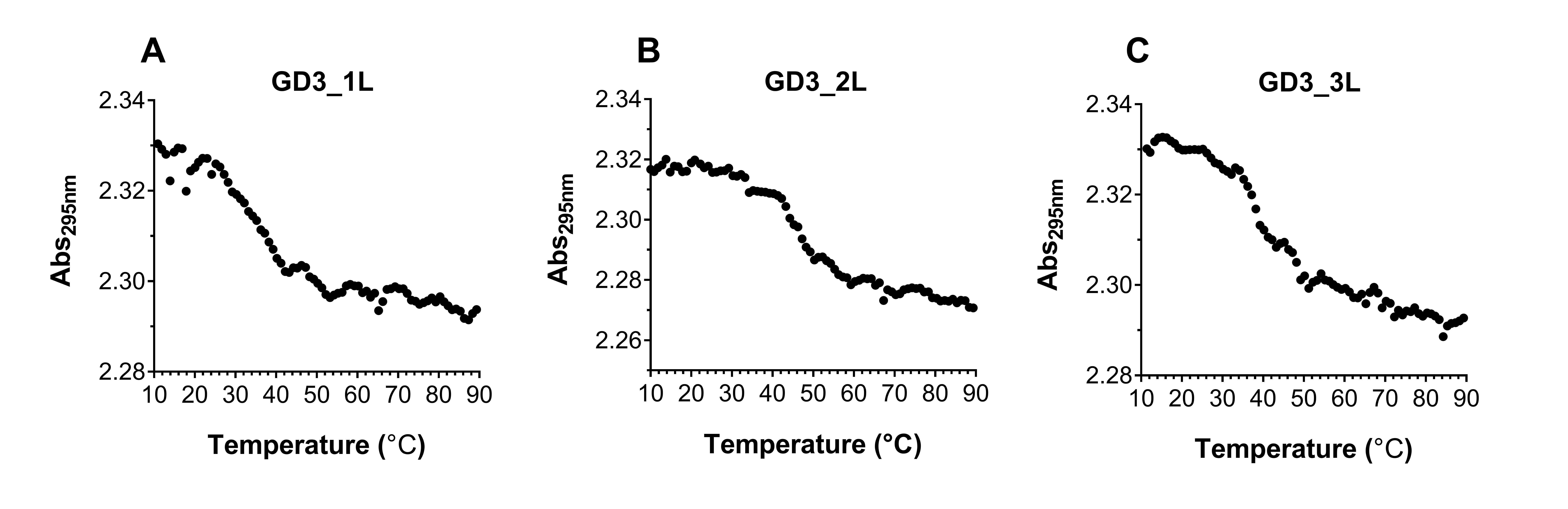
We elucidated the effect of the G4 CpG ODN sequence on the thermodynamic stability by evaluating the melting temperatures calculated from their UV melting curves at 295 nm (Figure 5). The melting temperature of GD3_1L is the lowest, indicating that GD3_1L is less stable than the other ODNs. On the other hand, the melting temperature of GD3_2L is higher than GD3_3L, suggesting that the thermodynamic stability of GD3_2L is higher than GD3_3L (Table 2). In addition, the decrease in the UV absorbance at 295 nm correlated to temperature indicates that all ODNs form G4 structures.
We analyzed the transcription levels of IL-6 and IL-12 produced in mouse macrophage-like RAW264 cells to evaluate the effects of the long loop position on the immunostimulatory activity induced by G4 CpG ODN. Figure 6 shows that RAW-264 cells exposed to the examined G4 CpG ODNs induced cytokine production at significantly higher levels than those exposed to the negative control, D-PBS, suggesting that all examined G4 CpG ODNs exhibit immunostimulatory activity. Furthermore, GD3_1L induced a higher IL-6 and IL-12 production than GD3_2L and GD3_3L, implying that GD3_1L has a higher immunostimulatory activity than GD3_2L and GD3_3L. Additionally, GD3_2L stimulated a higher cytokine production than GD3_3L.
Discussion
To explore the impact of loop permutation on the immunostimulatory activity of GD3_2L, we exchanged the sequence of the second loop with the first and the third loop to obtain GD3_1L and GD3_3L, respectively, as shown in Figure 2. The obtained ODN sequences differ in the sequential order of their three loops but have identical G-tracts, loop length, and loop composition.
Some studies reported that loop permutation does not stimulate the topology change in G4 structures. Fox et al. showed that despite the difference in the sequential orders of T and TTTT loops, d[(TG3)2T4(G3T)2] and d[TG3T4(G3T)3] adopt identical parallel topologies9. This adoption occurs due to two single-nucleotide loops in the sequence, which force the structure into a parallel fold, regardless of the remaining loop’s length10.
This study revealed the significant role of loop permutation in regulating the folding patterns of G4 CpG ODN. The exchange of the short first loop and the long second loop of GD3_2L leads to a change in the G4 topology from hybrid to parallel (Figure 4) and results in a decrease in the compactness and thermodynamic stability of GD3_2L (Figure 5 and Table 1). This shift in topology suggests that putting CpG motifs into the first loop is not preferable to form a stable G4 structure. Our results are compatible with the findings of Cheng et al., who reported that G4 with a longer central loop tends to form a stable non-parallel topology, and sequences with a short central loop have a high tendency to form a parallel topology with lower stability8. Meanwhile, swapping the order of the long second loop and the short third loop of GD3_2L allows the formation of a stable hybrid G4. Presumably, a long linker sequence at the first loop but not at the third loop enables the ODN to form a parallel G4 structure, which coheres with the observation of Cheng et al., who evaluated the impact of loop position exchange on the G4 folding11. They designed a set of ODNs having identical nucleotide compositions, loop length, loop composition, G-tracts, and flanking sequences except for the sequential order of loops11. The ODNs contain two TT loops and one TTA loop in the human telomeric sequence11. The ODN d[TT G3(TTG3)2TTAG3A], containing TTA sequence in the third loop, forms a non-parallel (hybrid or anti-parallel) G-quadruplex, while d[TTG3TTAG3(TTG3)2A], containing TTA in the first loop, forms a parallel G411. In addition, there is a difference between our study and the previous study in terms of the topology of the ODNs with the center loop longer than two external loops. The human telomeric sequence-based ODN with “TTA” in the central loop and “TT” in two external loops form parallel G411, while GD3_2L forms hybrid G4. This formation is perhaps attributable to the difference in the lengths of the central loop.
Loop transposition leads to a change in the biological function of G4 ODNs. For example, the 22-mer sequence containing the CpG motif of GD3_2L translocation from the second loop to the first loop correlates with an increase in cytokine induction. The higher cytokine induction of GD3_1L compared with GD3_2L may be attributed to the less compact structure of GD3_1L, as indicated by the PAGE results (Figure 4). Literature recognizes that a CpG ODN may adopt a bent conformation when it binds to both protomers of TLR9, resulting in TLR9 stimulation12. The flexibility of CpG motifs in higher-order structures has been demonstrated to play a vital role in TLR9 recognition and the subsequent immunostimulatory activities13. The looseness in the structure of GD3_1L reduces the rigidity of CpG motifs, enabling the CpG motifs to have more chances to interact and facilitate easier interaction with TLR9.
Meanwhile, swapping the second loop and the third loop of GD3_2L results in a decrease in the IL-6 and IL-12 induction. The structure of GD3_3L is as compact as GD3_2L’s structure (Figure 4), but GD3_3L is less effective than GD3_2L in TLR9 activation. Previously, we reported that inserting CpG motifs into different loops of RE31, a sequence forming anti-parallel G4 structure, leads to different immunostimulatory activities. Inserting the CpG motif into the first or third loop of RE31 preserves the anti-parallel structure of the wild-type, but the RE31 variant with CpG motifs in the third loop exhibits a significantly lower cytokine induction than the one with CpG motifs in the first loop14. The CpG motif located in the third loop of RE31 interacts with poly-guanine near the 3'-terminal and consequently forms a weaker interaction with TLR914. Although additional analyses should examine the structure of the examined G4 CpG ODNs and the interaction between TLR9 and G4 CpG ODNs, our results suggest that the factor directly influencing the immunostimulatory properties is not topology but rather the conformation of CpG motifs in G4.
Conclusions
The results of this study indicated that increasing the immunostimulatory properties of GD3_2L is possible by changing the sequential order of CpG motifs from the second to the first loop. This increase in efficacy may result from decreased structural compactness, which facilitates TLR9-CpG motif interaction. In addition, GD3_2L and GD3_3L have similar topology and structural compactness, but GD3_3L is considerably less effective than GD3_2L, suggesting that the G4 topology does not directly impact the immunostimulatory properties. The crucial factor determining the efficiency of G4 CpG ODNs may be the conformation of CpG motifs in the loop, regulated by several factors, including loop permutation.
Abbreviations
CD: circular dichroism, CpG: cytosine−phosphate− guanine, G4 CpG ODNs: G-quadruplex-based CpG ODNs, G4: G-quadruplex, IFN-β: interferon-β, IL-12: interleukin-12, IL-6: interleukin-6, ODNs: oligodeoxynucleotides, PAGE: polyacrylamide gel electrophoresis, RT-qPCR: reverse transcription quantitative-polymerase chain reaction, TLR9: Toll-like receptor 9, UV: ultraviolet
Acknowledgments
This research is funded by University of Science, VNU-HCM under grant number T2022-35. We are deeply grateful to M. Takeuchi, A. Junko, A. Nagai, H. Sanematsu for their technical suggestions for the CD and UV analyses. This work was carried out using the experimental equipment at the NIMS Molecule and Material Synthesis platform, supported by the Nanotechnology Platform Program of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, with payment of user fees.
Author’s contributions
All authors read and approved the final manuscript.
Funding
This work was supported by the Japan Society for the Promotion of Science Kakenhi grant number (17KK0122, 18K04858, and 21K19057) and University of Science, Vietnam National University, Ho Chi Minh City, Vietnam (T2022-35).
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
-
Wang
N.,
Chen
M.,
Wang
T.,
Liposomes used as a vaccine adjuvant-delivery system: from basics to clinical immunization. Journal of Controlled Release.
2019;
303
:
130-50
.
View Article PubMed Google Scholar -
Campbell
J.D.,
Vaccine Adjuvants. 2017;1494:15–27. Available from: http://link.springer.com/10.1007/978-1-4939-6445-1.
.
-
Meng
W.,
Yamazaki
T.,
Nishida
Y.,
Hanagata
N.,
Nuclease-resistant immunostimulatory phosphodiester CpG oligodeoxynucleotides as human Toll-like receptor 9 agonists. BMC Biotechnology.
2011;
11
(1)
:
88
.
View Article PubMed Google Scholar -
Tu
A.T.,
Hoshi
K.,
Ikebukuro
K.,
Hanagata
N.,
Yamazaki
T.,
Monomeric
G.,
Monomeric G-Quadruplex-Based CpG Oligodeoxynucleotides as Potent Toll-Like Receptor 9 Agonists. Biomacromolecules.
2020;
21
(9)
:
3644-57
.
View Article PubMed Google Scholar -
Hoshi
K.,
Yamazaki
T.,
Sugiyama
Y.,
Tsukakoshi
K.,
Tsugawa
W.,
Sode
K.,
G-quadruplex structure improves the immunostimulatory effects of CpG oligonucleotides. Nucleic Acid Therapeutics.
2019;
29
(4)
:
224-9
.
View Article PubMed Google Scholar -
Tu
A.T.T.,
Hoshi
K.,
Shobo
M.,
Yamazaki
T.,
G-quadruplex-based CpG oligodeoxynucleotide/DOTAP complex strongly stimulates immunity in CpG motif-specific and loop-length-dependent manners. Nanomedicine.
2022;
40
:
102508
.
View Article PubMed Google Scholar -
Tu
A.T.T.,
Hoshi
K.,
Ma
Y.,
Oyama
T.,
Suzuki
S.,
Tsukakoshi
K.,
Nagasawa
K.,
Ikebukuro
K.,
Yamazaki
T.,
Effects of G-Quadruplex Ligands on the Topology, Stability, and Immunostimulatory Properties of G-Quadruplex-Based CpG Oligodeoxynucleotides. ACS Chemical Biology.
2022;
17
(7)
:
1703-13
.
View Article PubMed Google Scholar -
Cheng
M.,
Cheng
Y.,
Hao
J.,
Jia
G.,
Zhou
J.,
Mergny
J.L.,
Loop permutation affects the topology and stability of G-quadruplexes. Nucleic Acids Research.
2018;
46
(18)
:
9264-75
.
View Article PubMed Google Scholar -
Rachwal
P.A.,
Findlow
I.S.,
Werner
J.M.,
Brown
T.,
Fox
K.R.,
Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Research.
2007;
35
(12)
:
4214-22
.
View Article PubMed Google Scholar -
Bugaut
A.,
Balasubramanian
S.,
A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry.
2008;
47
(2)
:
689-97
.
View Article PubMed Google Scholar -
Cheng
M.,
Zhou
J.,
Jia
G.,
Ai
X.,
Mergny
J.L.,
Li
C.,
Relations between the loop transposition of DNA G-quadruplex and the catalytic function of DNAzyme. Biochimica et Biophysica Acta. G, General Subjects.
2017;
1861
(8)
:
1913-20
.
View Article PubMed Google Scholar -
Ohto
U.,
Shibata
T.,
Tanji
H.,
Ishida
H.,
Krayukhina
E.,
Uchiyama
S.,
Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature.
2015;
520
(7549)
:
702-5
.
View Article PubMed Google Scholar -
Sanada
Y.,
Shiomi
T.,
Okobira
T.,
Tan
M.,
Nishikawa
M.,
Akiba
I.,
Polypod-shaped DNAs: small-angle X-ray scattering and immunostimulatory activity. Langmuir.
2016;
32
(15)
:
3760-5
.
View Article PubMed Google Scholar -
Safitri
F.A.,
Tu
A.T.,
Hoshi
K.,
Shobo
M.,
Zhao
D.,
Witarto
A.B.,
Enhancement of the Immunostimulatory Effect of Phosphodiester CpG Oligodeoxynucleotides by an Antiparallel Guanine-Quadruplex Structural Scaffold. Biomolecules.
2021;
11
(11)
:
1617
.
View Article PubMed Google Scholar
Comments
Article Details
Volume & Issue : Vol 9 No 11 (2022)
Page No.: 5410-5417
Published on: 2022-11-30
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 4249 times
- PDF downloaded - 1343 times
- XML downloaded - 0 times
 Biomedpress
Biomedpress