Abstract
Introduction: Proteases are enzymes that catalyze the hydrolysis of peptide bonds and play an important role in almost all biological processes. However, excessive protein proteolysis can be implicated in several diseases, such as cancer, as well as cardiovascular, inflammatory, neurodegenerative, bacterial, viral and parasitic diseases. In these cases, protease inhibitors can be used as one of versatile tools for regulating proteolytic activity of target proteases as well as therapeutic applications. In this study, we expressed and characterized a new serine protease inhibitory protein (PI-QT) from the metagenome of sponge-associated microorganisms in Escherichia coli.
Methods: The gene PI-QT encoding for a new serine protease inhibitory protein was expressed in E. coli BL21(DE3). In addition, the expressed protein was purified and characterized.
Results: Optimization of expression of the recombinant protein PI-QT in E. coli showed that suitable conditions for expression of the protein were pre-induction cell density (OD600) of 0.6 - 0.7, IPTG concentration of 1 mM and temperature of 25oC. The protease inhibitory protein was also purified and identified by mass spectrometry LC-MS/MS. The recombinant protein showed inhibitory activity against trypsin anda-chymotrypsin with activity values of 97526 U/mg and 41714 U/mg, respectively. Maximum activity of the protease inhibitory protein was obtained at pH 7 and temperature 20-35oC. The inhibitor was stable over pH 4-9 and up to temperature 50oC. Addition of Zn2+, Mg2+ and Ca2+ enhanced inhibitory activity, whereas other metal ions, surfactants and oxidants reduced inhibitory activity of the protease inhibitor.
Conclusion: The recombinant protein PI-QT is a potential protease inhibitor for therapeutic applications.
Introduction
Proteases are enzymes that catalyze the hydrolysis of peptide bonds and play an important role in almost all biological processes; however, their uncontrolled activity often leads to diseases. Excessive protein proteolysis can relate to several diseases, including cancer, cardiovascular, inflammatory, neurodegenerative, bacterial, viral and parasitic diseases1. In these cases, protease inhibitors (PIs) can be used as one of the versatile tools for regulating proteolytic activity of target proteases2. To date, enzyme inhibitors have received increasing attention, not only for discovery of their structures and action mechanisms but also for potential application in different fields3, 1.
Marine sponges are known to harbor diverse microbial communities6, 5, 4 and represent a prolific source of natural products from their associated microorganisms10, 9, 8, 7. Recent studies have shown that many potential protease inhibitors have been isolated from sponge-associated microorganisms14, 13, 12, 11; however, the exploration and exploitation potential PIs from microorganisms are still a big challenge because almost all microorganisms are resistant to cultivation in laboratory conditions, especially symbiotic microbes. Fortunately, new approaches (e.g., metagenomics) provide powerful tools for predicting, detecting and expressing novel bioactive genes from non-cultured microorganisms19, 18, 17, 16, 15. This opens new avenues for discovering new bioactive compounds, including PIs, in the future.
In order to meet the demands of finding novel and potential PIs from the marine environment, especially PIs from sponge-associated microorganisms, we optimized the expression and characterization of a new serine protease inhibitory protein PI-QT in an E. coli expression system.
Methods
Materials
The gene PI-QT is a new gene encoding a serine protease inhibitor from the metagenome of marine sponge QT collected from Quang Tri, Vietnam. It was synthesized and inserted into cloning vector pUC57 (GenScript, Piscataway, NJ, USA). The sequence of gene PI-QT was deposited in the National Center for Biotechnology Information (NCBI) database with the accession number MK359987.
Sequence analysis, multiple sequence alignment, and phylogeny analysis
The PI-QT sequence was compared with other known sequences in the NCBI database using the Blast program. The open reading frame (ORF) of PI-QT was determined using ORF Finder program (https://www.ncbi.nlm.nih.gov/orffinder/). The molecular weight and pI of the deduced protein PI-QT were examined using the Compute pI/Mw tool of the Expasy server (http://web.expasy.org/compute_pi/). Multiple sequence alignments were performed using the ClustalW algorithm in the MEGA 7.0 software (http://megasoftware.net). A phylogenetic tree was constructed by the neighbor-joining (NJ) method and support of a bootstrap analysis with 1,000 replications implemented in the MEGA 7.0 software. Protein structure model of PI-QT was predicted using SWISS-MODEL program (https://swissmodel.expasy.org/) and (PS)2-v2: protein structure prediction server (http://ps2.life.nctu.edu.tw/).
Construction of expression vector pET-32a(+)/PI-QT
The vector pUC57 (Thermo Fisher Scientific, Waltham, MA, USA) containing the gene PI-QT and expression vector pET-32a(+) (Invitrogen, Carlsbad, CA, USA) were digested with restriction enzymes EcoRI and NotI (Fermentas, Vilnius, Lithuania). The gene PI-QT was then purified and inserted into expression vector pET-32a(+) using enzyme T4 ligase (Thermo Fisher Scientific).
Optimization of the expression of recombinant protein PI-QT
The recombinant vector pET-32a(+)/PI-QT was transformed into E. coli strain BL21(DE3) (Invitrogen) by the heat shock method. The transformed cells were plated on LB plates containing 50 µg/mL ampicillin (LB/amp) (Sigma-Aldrich, St. Louis, MO, USA) and incubated overnight at 37°C. Expression of the recombinant protein PI-QT was optimized using different conditions such as temperature (20, 25, 30, 37oC), IPTG (Sigma-Aldrich, St Louis, MO, USA) concentration (0.1, 0.5, 1.0, 1.5 mM), and pre-induction cell density (OD600 = 0.4, 0.5, 0.6, 0.7).
Determination of protein in soluble and insoluble fractions
Cell culture was centrifuged at 10,000 rpm for 15 min and then supernatant removed. The cells were resuspended in TE buffer (Tris 20mM, EDTA 10mM, PM F 0.05mM) and incubated at -75oC for 1 hr, then thawed at 50oC for 30 min, and sonicated on ice with a Misonix Ultrasonic Liquid Processors (company name & location). The solution was centrifuged at 13,000 rpm for 15 min, then the supernatant (soluble fraction) collected. The pellet was resuspended in TE buffer to the original volume (insoluble fraction). Next, SDS loading buffer was added to the pre-treatment culture (total protein), soluble protein fraction and insoluble protein fraction , and then the proteins were denatured at 100oC for 10 min. The expression of the protein was checked on 12.6% SDS-PAGE gel.
Western blot analysis
After electrophoresis on SDS-PAGE gel, the recombinant protein was transferred to polyvinylidene difluoride (PVDF) membrane. The membrane was then blocked with TBS (containing 5% skimmed milk) and incubated with anti-TRx antibody (Sigma-Aldrich, St Louis, MO, USA) at a dilution of 1:1000 for overnight at 4 oC. The membrane was washed 4 times with TBS washing buffer and incubated with anti-mouse IgG secondary antibody at a dilution of 1:5000 for 1 hr at room temperature. The membrane was washed with TBS washing buffer and visualized by addition of ρ-nitro blue tetrazolium chloride 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) solution.
Purification of protein PI-QT
Purification of the recombinant protein PI-QT was performed by chromatography on Ni-NTA affinity chromatography column and then eluting with imidazole at concentrations of 100, 300, and 500 mM. The purified protein was removed from salts by dialysis tube (SnakeSkinTM Dialysis tubing) (Thermo Fisher Scientific) in TBS buffer (50 mM Tris HCl, 50 mM NaCl, pH 7.4) at 4 oC for 24 h. Next, the recombinant protein was removed from the TRx-tag fusion using Thrombin kit (Novagen, Darmstadt, Hesse, Germany).
Protease inhibitory assay
Trypsin inhibition assay
The purified protein (50 μl of 0.05 mg/ml) was added to a mixture of 50 μl of trypsin (Sigma-Aldrich) solution (0.05 mg trypsin/ml of 0.05 M Tris-HCl) and 100 μl of 0.05 M Tris-HCl, pH 8.0, containing 0.03 M CaCl2. The mixture was incubated at 37°C for 10 min, then 1.0 ml of 0.8 mM BapNA (Sigma-Aldrich) solution was added and incubated at 37°C for 10 min. The reaction was stopped by adding 20 µl of 30 % (v/v) glacial acetic acid. Subsequently, the solution was centrifuged at 10,000 rpm for 15 min and the absorbance were measured at 410 nm against appropriate blanks.
ɑ- Chymotrypsin inhibition assay
The purified protein (50 μl of 0.05 mg/ml) was added to a mixture of 50 μl of ɑ-chymotrypsin (Sigma-Aldrich) solution (0.05 mg ɑ-chymotrypsin/ml of 0.05 M Tris-HCl) and 100 μl of 0.05 M Tris-HCl, pH 8.0, containing 0.03 M CaCl2. The mixture was incubated at 37°C for 10 min, then 1.0 ml of 0.86 mM BTpNA (Sigma-Aldrich) solution was added and incubated at 37°C for 10 min. The reaction was stopped by adding 20 µl of 30 % (v/v) glacial acetic acid. Subsequently, the solution was centrifuged at 10,000 rpm for 15 min and the absorbance were measured at 410 nm against appropriate blanks.
Bowman-Birk Inhibitor (BBI) (Sigma-Aldrich) was used as a positive control for trypsin and ɑ-chymotrypsin inhibition assays.
Identification of the recombinant protein
Tryptic digestion in gel
The protein band from SDS-PAGE was excised, destained and dehydrated with 50% acetonitrile. The protein was reduced with DTT (65 mM) and subsequently alkylated with IAA (55 mM). The peptides were desalted using Zip-Tips (Millipore, Bedford, MA) according to the manufacturer’s instructions. The purified peptides were collected for analysis by LC-MS/MS.
Identification of the recombinant protein by shotgun proteomics
NanoLC-nanoESI-MS/MS analysis was performed on a nanoAcquity system (Waters, Milford, MA, USA) connected to an OrbitrapVelos hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a PicoViewnanospray interface (New Objective, Woburn, MA, USA). Peptide mixtures were loaded onto a 75 μm ID, 25 cm length C18 BEH column (Waters, Milford, MA, USA), packed with 1.7 μm particles with a pore size of 130 Å, and were then separated using a segmented gradient in 60 min from 5% to 40% solvent B (acetonitrile with 0.1% formic acid) at a flow rate of 300 nl/min and a column temperature of 35°C. Solvent A was 0.1% formic acid in water. The mass spectrometer was operated in the data-dependent mode. After acquisition of spectra, the proteins and peptides were identified by the PEAKS software (Bioinformatics Solutions Inc., Ontario, Canada).
Characterization of protease inhibitor PI-QT
Effect of pH and temperature on activity of the protease inhibitor were determined by performing protease inhibitor assay at pH 7 after incubating the purified protease inhibitor in buffers of different pH (2-12) for 24 h and at different temperatures (30◦C to 70◦C) for 1 h.
Effect of metal ions, surfactants, and oxidizing agents on activity of the protease inhibitor were evaluated after incubating the protease inhibitor with 1 mM concentrations of metal salts (MgSO4, CuSO4, ZnSO4, CaCl2, MnCl2, FeCl2, NaCl), 1% (v/v) of surfactants (Tween 20, Tween 80 and Triton X 100), and oxidants (H2O2 and DMSO) for 30 min at 37 ◦C.
Statistical Analysis
The assays were performed in triplicate and expressed as the mean ± standard error of the mean (SEM). The statistical analysis was performed by t-test and one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests using the SPSS v.22 (SPSS Inc, Chicago, IL, USA). The results were considered to be significant at P < 0.05.
Results
Amino acid sequence and phylogeny analysis of protein PI-QT
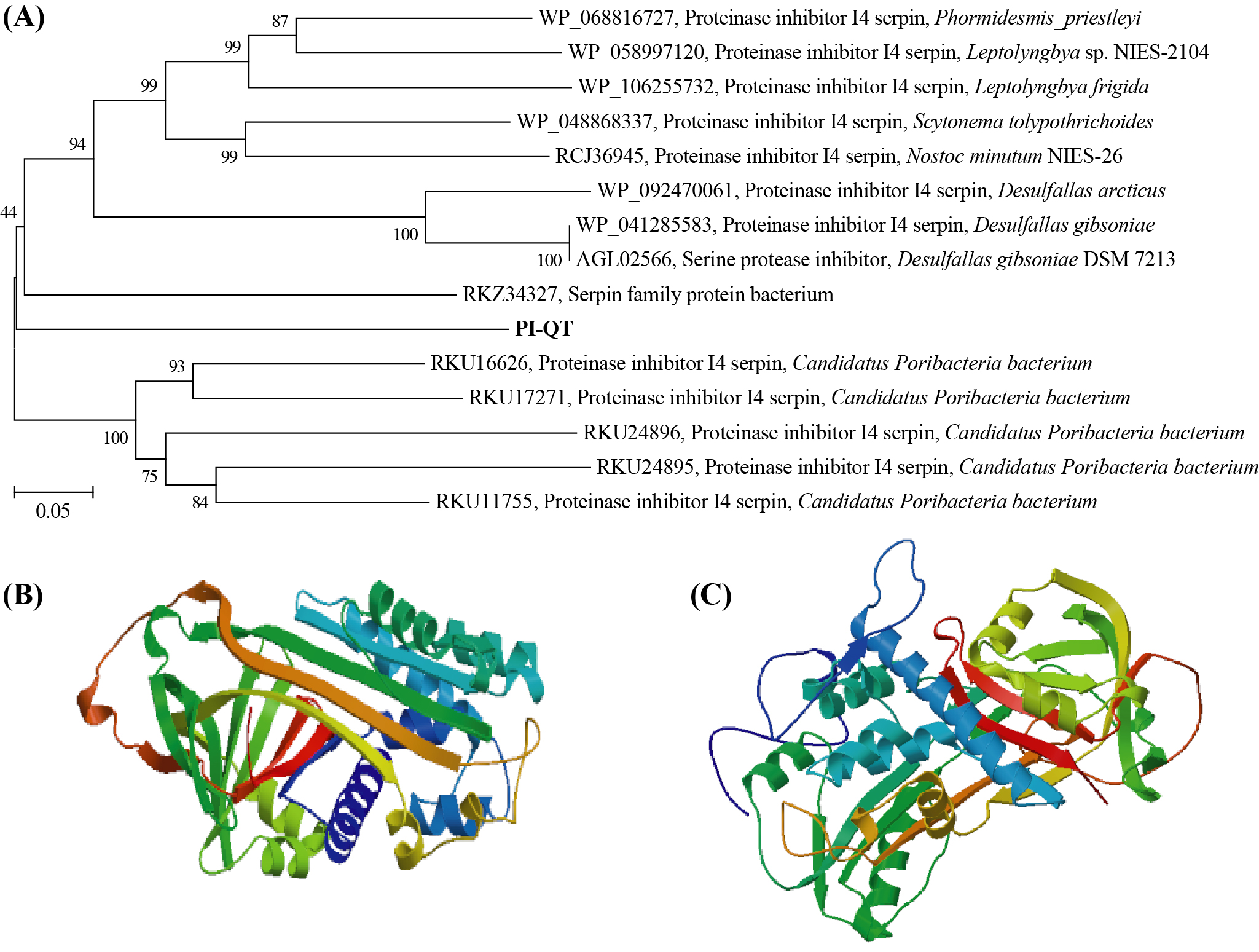
The gene PI-QT was 1,287 bp in length and had an open reading frame of 429 amino acid with a calculated molecular mass of about 50 kDa and a theoretical isoelectric point of 4.56. Comparison of the deduced amino acid sequence of PI-QT with the sequences in the NCBI database showed that protein PI-QT was most similar with serpins with similarities <55%. Multiple alignments of the deduced amino acids of PI-QT with the most homologous serpins in NCBI database showed that the deduced peptide PI-QT shared conserved active site residues with microbial serpin members (Figure 1). The phylogenetic tree based on the neighbor-joining method (Figure 2 A) located the protein PI-QT between two microbial serpin clades: one serpin clade from a candidate phylum of bacteria (Poribacteria) originally identified in the microbiome of marine sponges and another serpin clade from bacterial phyla Firmicutes and Cyanobacteria. Based on the database comparison, the protein PI-QT was considered as a new microbial serpin. The protein structure of PI-QT was predicted by SWISS-MODEL (Figure 2 B) and (PS)2-v2 model (Figure 2 C).
Construction of the expression vector pET-32a(+)/PI-QT
The vectors pUC57 containing gene PI-QT and the expression vector pET-32a(+) were digested with EcoRI and NotI (Supplementary 1A). The gene PI-QT was then inserted into the opened expression vector pET-32a(+) using enzyme T4 ligase, transformed into E. coli strain TOP10F', and plated on LB plate supplemented with ampicillin (50 µg/mL). Agarose gel analysis of the plasmids isolated from colonies and cut with enzymes EcoRI and NotI (Supplementary 1B) showed bands of ~1.3 kb (corresponding to size of gene PI-QT) and bands of 5.9 kb (corresponding to size of vector pET-32a(+)). In addition, the sequencing results of the plasmid showed that the gene sequence inserted into the vector was identical to the sequence of the designed gene PI-QT (data not shown), thereby demonstrating that gene PI-QT was inserted successfully into expression vectors pET-32a(+) to form the recombinant vector pET-32a(+)/PI-QT.
Expression of recombinant protein in E.coli strain BL21(DE3)
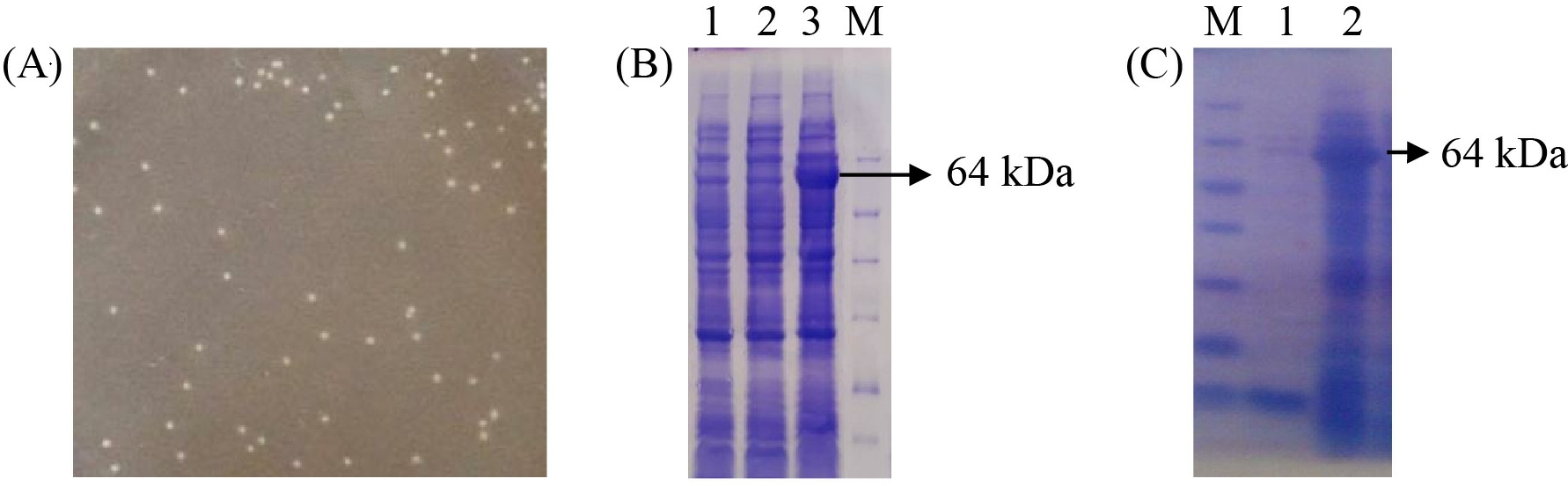
The recombinant vector pET-32a(+)/PI-QT was transformed into E.coli BL21(DE3) by heat shock and incubated on LB/amp medium overnight. Therefore, white colonies (Figure 3 A) were incubated in LB/amp broth until OD600 was about 0.8 - 1.0 IPTG was then added in culture and incubated at 37oC for 4 h. SDS-PAGE gel analysis of protein expression profile showed the presence of an overexpressed protein band of 64 kDa (Figure 3 B, lane 3), corresponding to the size of recombinant protein PI-QT with Trx-tag fusion, whereas this foreign protein band was not present in negative control sample (Figure 3 B, lane 1) containing only vector pET32a(+) and samples containing the recombinant vectors but not induced with IPTG (Figure 3 B, lane 2). However, the recombinant protein was expressed mainly in the insoluble fraction (Figure 3 C, lane 2). Therefore, optimization of the expression conditions is necessary to increase the amount of the recombinant protein in the soluble fraction.
Optimization of recombinant protein expression
Determination of suitable IPTG concentration for recombinant protein expression
Expression of the recombinant protein PI-QT was induced with different IPTG concentrations (0, 0.5, 1.0, and 1.5 mM). The SDS-PAGE gel analysis showed the recombinant protein was expressed at 1 mM IPTG, but not at low or high concentration of IPTG (0.5 mM and 1.5 mM). However, the expressed protein was still mainly present in the insoluble fraction (data not shown).
Determination of suitable temperature for recombinant protein expression
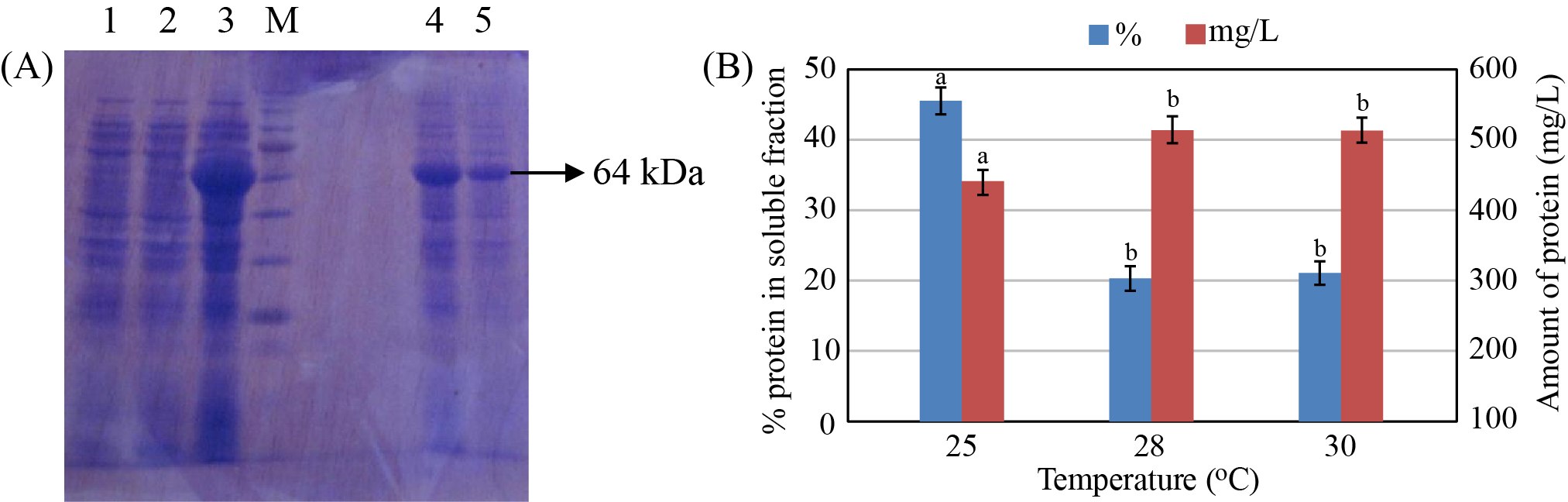
The expression of the recombinant protein was also investigated at different temperature (20, 25, 28, and 30 oC). The experimental results showed that the amount of recombinant protein in soluble fraction reached the highest value (423 mg/L with about 45% in the soluble fraction) when the recombinant protein was expressed at 25 oC (Figure 4), whereas the expression of recombinant protein was not observed at 20 oC (data not shown). At 28 oC and 30 oC, the amount of recombinant protein produced was higher; however, only a small amount of the recombinant protein was detected in the soluble fraction (only about 20% of total protein).
Determination of suitable cell density for recombinant protein expression
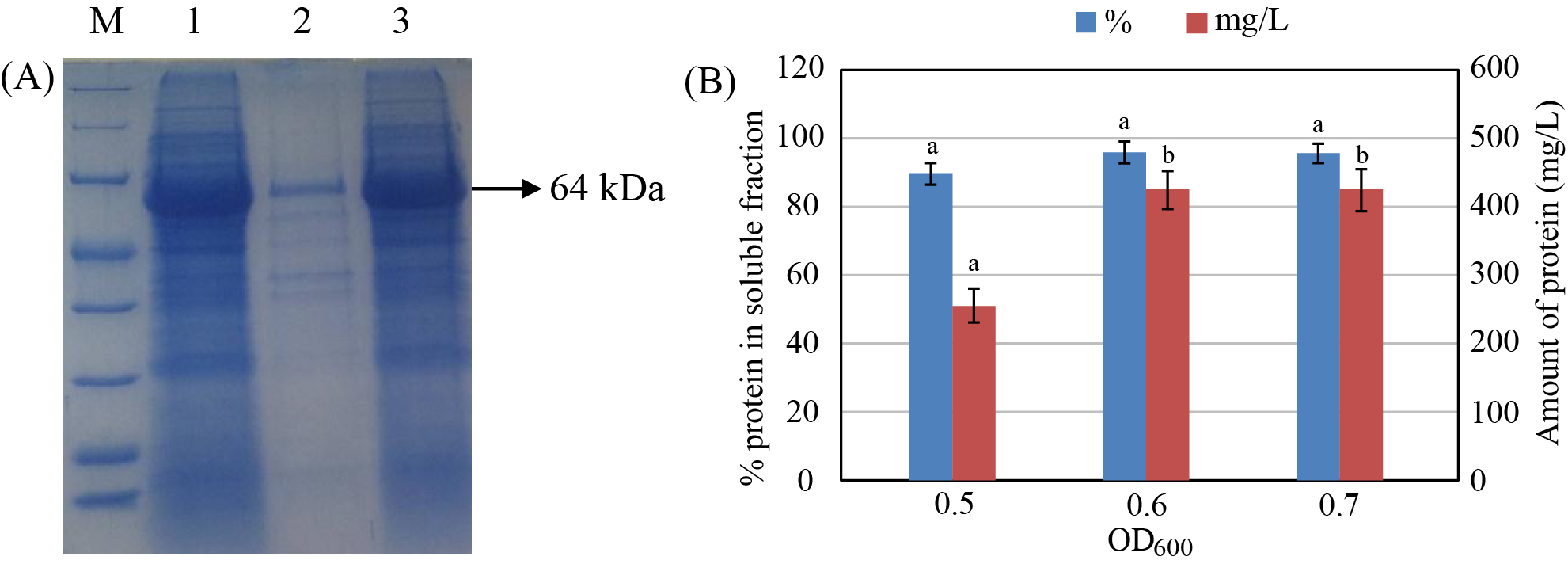
The expression of the recombinant protein at different pre-induction cell densities (OD600 = 0.4, 0.5, 0.6, and 0.7) was investigated (Figure 5). The results showed that expression of the protein was not observed at OD600 = 0.4. The total amount of produced protein and protein content in the soluble fraction were increased with increasing pre-induction cell density and reached the highest values at OD600 = 0.6 - 0.7 (409 mg/L with 90% in the soluble fraction).
Purification and identification of the recombinant protein
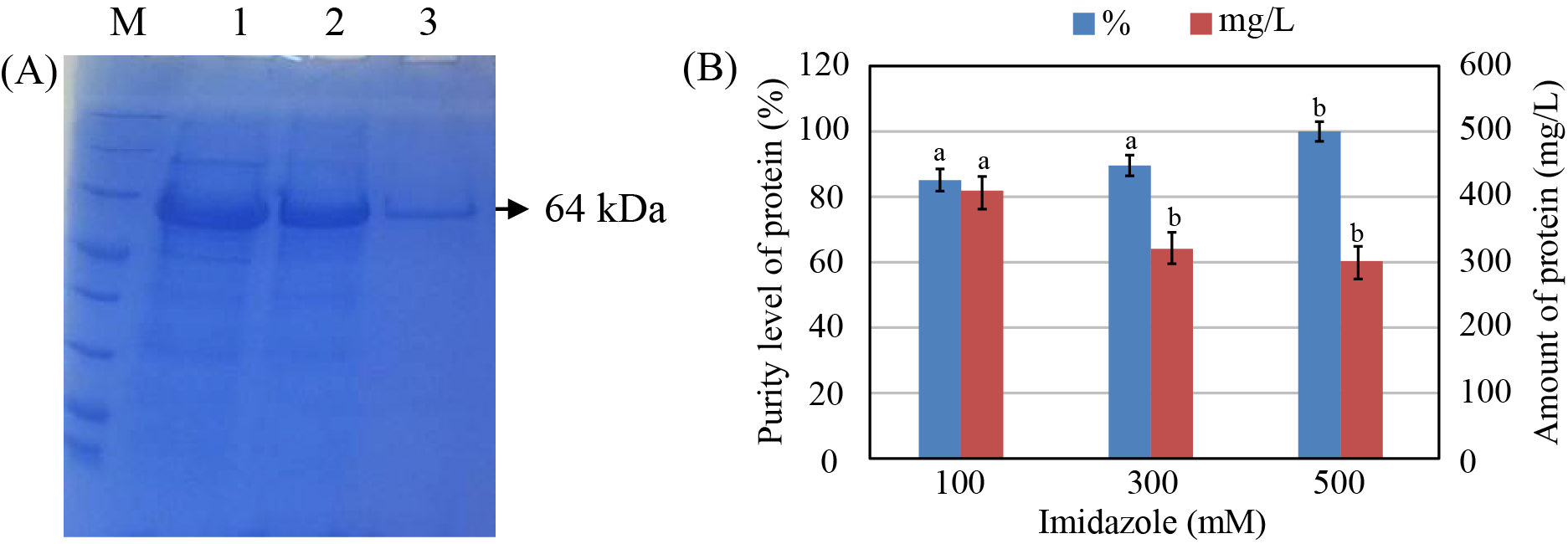
The purification of the recombinant protein by Ni-NTA affinity chromatography column showed that the recombinant protein was of high yield with imidazole concentrations of 100 and 300 mM; however, protein purity was low. In the case of imidazole concentration of 500 mM, the amount of the obtained protein was lower but had higher purity than those of the two above cases (Figure 6).
To confirm that the purified protein was the protein PI-QT, we performed Western lot assay using anti-Trx antibody. The Western lot analysis showed the presence of a protein band of 64 kDa on the hybrid membrane that was similar to the size of protein PI-QT with Trx-tag fusion (Figure 7 A). In addition, in order to remove the Trx-tag fusion from the recombinant protein PI-QT, the protein was cut off from Trx-tag by thrombin. SDS-PAGE gel analysis of the protein treated with thrombin showed 2 bands of 50 kDa and 14 kDa, which corresponded to the size of protein PI-QT and Trx-tag, respectively (Figure 7 B). This revealed that the Trx-tag fusion was removed successfully from the recombinant protein.
In order to identify the recombinant protein by mass spectrometry, the recombinant protein was cut, hydrolyzed, chromatographed, and analyzed for spectroscopic data (see methods). The LC-MS/MS spectroscopic data analysis of the recombinant protein (Supplementary 2 & 3) revealed that the sequence of the peptide fragments hydrolyzed and extracted from the recombinant protein was identical to the designed sequence. Since the protein was hydrolyzed into small peptides and only about 30% of peptides were recovered (Supplementary 3), it is therefore not possible to find similar polypeptides on available databases. However, analyses based on the protein database built from the designed sequence by the software PEAKS and of the recovered peptide sequences confirmed that the expressed and purified recombinant protein was the desired protein.
Activity and characterization of the recombinant protease inhibitor
The recombinant protein was evaluated for its protease inhibitory activity against trypsin and ɑ-chymotrypsin. Protease inhibitory assay showed that the recombinant protein exhibited inhibitory effects against trypsin and α-chymotrypsin with specific activities of 975 ± 26 U/mg and 417 ± 14 U/mg, respectively. Compared to protein PI-QT, BBI (positive control) showed better inhibitory effects against trypsin and α-chymotrypsin with specific activities of 3303 ± 66 U/mg and 1340 ± 58 U/mg, respectively.
The activity of the protease inhibitor peaked at pH 7 and still maintained more than 60% of its activity within pH 4-9. The activity of the protease inhibitor sharply declined at highly acidic (pH 3) and alkaline (pH 10) conditions (Figure 8 A). The obtained results also showed that the protease inhibitor was most active at temperatures of 20-35 oC, and still maintained more than 60% of its activity up to 50 oC. The protease inhibitor activity decreased rapidly at temperature >60 oC (Figure 8 B).
The experimental results showed that the presence of surfactants (Tween 20, Tween 80, and Triton X 100) led to decreases of activity of the protease inhibitor compared to the control (Figure 8 C). Furthermore, the activity reduction of the protease inhibitor was observed with the presence of oxidizing agents, H2O2 and DMSO (Figure 8 C).
Effects of metal ions on the protease inhibitor were observed in this study (Figure 8 D). The obtained results showed that the presence of Zn2+, Mg2+, and Ca2+ ions enhanced the activity of the protease inhibitor, whereas the presence of Cu2+, Mn2+, Fe2+, and Na+ ions had negative effects and did not support protease inhibitor activity compared to the control.
Discussion
Protease inhibitors play an important role in the regulation of protease activity and have been used as a potential tool in different fields. Indeed, the discovery and exploitation of novel PIs from different sources have garnered increased attention across scientific research areas. In the present study, we have expressed and characterized a new serine protease inhibitor protein (PI-QT) from the metagenome of sponge-associated microorganisms in E. coli. The homogenous serpins in the NCBI database with the protein PI-QT are from microorganisms, suggesting that the protein PI-QT is a microbial serpin. Interestingly, several homogeneous serpins with the protein PI-QT have been detected from sponge-associated bacteria (candidate phylum Poribacteria) based on metagenome data.
The protein PI-QT had an open reading frame of 429 amino acid with a calculated molecular mass of about 50 kDa. These values were consistent with the observed average molecular mass of most serpin proteins, ranging from 40 - 60 kDa with 330 - 500 amino acids20. The protein PI-QT exhibited inhibitory effects against trypsin and α-chymotrypsin with specific activities of 975 ± 26 and 417 ± 14 U/mg, respectively. The specific activity of the protein PI-QT was comparable with that of novel serpins reported in recent studies24, 23, 22, 21. For example, Jiang et al.21 have cloned and expressed a novel serpin (Spi1C) from metagenomic library of uncultured marine microorganisms; the Spi1C protein exhibited inhibitory effects against trypsin and α-chymotrypsin with specific values of 6940 and 3640 U/mg, respectively. Chan et al. 22 have purified a thermostable trypsin inhibitor from small pinto beans and reported that the purified protein showed inhibitory activity against trypsin with specific value of 2398 U/mg. Shamsi et al.23 have isolated and purified a novel Kunitz trypsin inhibitor (ASPI) from garlic Allium sativum; the ASPI protein showed inhibitory activity against trypsin with specific activity of 30376 U/mg. In another study, Mohan et al.24 purified and characterized a protease inhibitor from Capsicum frutescens with specific activity against trypsin of 6749 U/mg.
The effects of IPTG concentration, pre-induction cell density, and temperature on the expression of the protease inhibitor PI-QT were evaluated to investigate the suitable and optimal conditions for expression of the protease inhibitor PI-QT in E. coli. Previous studies have reported that the synthesis of recombinant protein by expression vector pET-32a(+) is controlled by the T7 promoter and that this promoter is induced by the presence of IPTG in the culture medium. The concentration of IPTG in culture medium can, therefore, influence the expression of recombinant protein. The transcription may be hindered and the amount of foreign protein produced may be reduced due to low concentrations of IPTG in medium, whereas high concentrations of IPTG can cause cytotoxicity and inhibit cell growth25. In addition, it has also been reported that the optimal temperature for growth of E. coli is 37 oC; however, this temperature is not suitable for producing foreign protein. The high temperature may result in eliminating plasmids. In addition, expression of protein at high temperatures can produce proteins with a tertiary structure, resulting in the loss of biological activity of proteins, whereas low expression temperature may reduce the cleavage of target protein by intracellular protease and significantly increase the amount of recombinant protein27, 26.
The experiments also showed that the protease inhibitor PI-QT were stable and maintained its activity within a wide range of pH and temperature. The stability of the protease inhibitor PI-QT within a wide range of pH and temperature is one of promising and interesting characteristic for its applications in biotechnological and pharmaceutical industries. Similar results were observed for many protease inhibitors of the Kunitz family in previous studies. The protease inhibitors in the Kunitz family are stable in wide pH ranges (pH 4-10), but sensitive to extreme pH conditions31, 30, 29, 28, 21. Under strongly acidic or alkaline conditions, the proteinaceous inhibitors are denatured and then lose their activity partially or completely. In addition, high temperature can affect the intramolecular disulfide bridges which are presumably responsible for the functional stability of Kunitz type protease inhibitors32.
The negative effects of surfactant and oxidizing agents on the activity of the protease inhibitor PI-QT were also observed in this study. The effects of surfactants on the activity of protease inhibitors could be attributed to a reduction of hydrophobic interactions, whereas effects of oxidizing agents on the activity of protease inhibitors may be attributed to the probable oxidation of the amino acid methionine at the reactive site of the inhibitors33. Furthermore, metal ions can reduce the activity of the protease inhibitor PI-QT, with the exception of some metal ions, such as Mg2+, Ca2+ and Zn2+.
Our study, together with previous studies, reveal that exploration of new bioactive compounds by expression of metagenome-based screening genes can be a versatile and potential strategy for mining new natural products from the marine environment as well as other environments17, 16, 15.
Conclusions
A new gene encoding a serine protease inhibitor PI-QT screened from the metagenome of sponge-associated microorganisms was optimized and expressed successfully in E. coli. The recombinant protein had a molecular mass of about 50 kDa. The Western lot assay and spectroscopic data analyses co firmed expression of the recombinant protein PI-QT. The recombinant protein exhibited inhibitory activity against trypsin and α-chymotrypsin with 975 ± 26 U/mg and 417 ± 14 U/mg, respectively. The protease inhibitor was stable within a wide range of pH and temperature. This study has shown that expression of recombinant protein from metagenome-based screening genes is a promising tool for the discovery and potential use of new bioactive compounds.
ABBREVIATIONS
Amp: Ampicillin
BApNA: Nα-Benzoyl-L-arginine 4-nitroanilide
BTpNA: N-Benzoyl-L-tyrosine p-nitroanilide
DMSO: Dimethyl sulfoxide
DTT: Dithiothreitol
EDTA: Ethylenediaminetetraacetic acid
IAA: Iodoacetamide
IPTG: Isopropyl β-D-1-thiogalactopyranoside
LB: Luria-Bertani broth
LC-MS/MS: Liquid chromatography-tandem mass spectrometry
NCBI: National Center for Biotechnology Information
OD: Optical density
PI: Protease inhibitor
PMSF: Phenylmethylsulfonyl fluoride
PVDF: polyvinylidene difluoride membrane
SDS: Sodium dodecyl sulfate
SDS-PAGE: Sodium dodecyl sulfate-Polyacrylamide gel electrophoresis
TBS: Tris-buffered saline
TE: Tris-EDTA
Competing Interests
The authors declare no conflict of interest.
Authors' Contributions
Conceived and designed the experiments: Tran Thi Hong, Ton That Huu Dat, Nguyen Thi Kim Cuc, Pham Viet Cuong, Le Minh Bui. Performed the experiments: Tran Thi Hong, Ton That Huu Dat, Nguyen Phuong Hoa, Tran Thi Kim Dung, Vu Thi Thu Huyen. Analyzed the data: Tran Thi Hong, Ton That Huu Dat. Contributed reagents/materials/analysis tools: Le Minh Bui, Nguyen Thi Kim Cuc, Pham Viet Cuong. Wrote and revised the manuscript: all authors. All authors read and approved the final manuscript.
Acknowledgements
LTQ-Orbitrap data and additional technical assistance were supported by the Academia Sinica Common Mass Spectrometry Facilities located at the Institute of Biological Chemistry, Taiwan.
FUNDING
This work was financially supported by Vietnamese Ministry of Science and Technology under grant number: ĐTĐLCN.17/14.
References
-
Saboti\vc
J.,
Kos
J.,
Microbial and fungal protease inhibitors\textemdashcurrent and potential applications. Appl Microbiol Biotechnol.
2012;
93
(4)
:
1351-75
.
View Article PubMed Google Scholar -
Shamsi
T.N.,
Parveen
R.,
Fatima
S.,
Characterization, biomedical and agricultural applications of protease inhibitors: A review. Int J Biol Macromol.
2016;
91
:
1120-33
.
View Article PubMed Google Scholar -
Ryan
B.J.,
Henehan
G.T.,
Overview of Approaches to Preventing and Avoiding Proteolysis During Expression and Purification of Proteins. Curr Protoc Protein Sci.
2013;
71
(1)
:
5.25.1-5.25.7
.
View Article Google Scholar -
Cuc
N.T.K.,
Dat
T.T.H.,
Hong
T.T.,
Cuong
P.V.,
Phylogenetic diversity of microorganisms associated with three marine sponges from Mien Trung sea of Vietnam. Vietnam Journal of Science and Technology..
2017;
55
(2)
:
168
.
View Article Google Scholar -
Dat
T.T.H.,
Cuong
P.V.,
Dung
T.T.K.,
Huyen
V.T.T.,
Cuc
N.T.K.,
Biodiversity assessment of microorganisms associated with two marine sponges (Haliclona oculata and Amphius huxleyi) collected at the Lang Co bay of Vietnam. J Mar Sci Technol.
2018;
18
(3)
:
286-95
.
View Article Google Scholar -
Dat
T.T.H.,
Steinert
G.,
Cuc
N. Thi Kim,
Smidt
H.,
Sipkema
D.,
Archaeal and bacterial diversity and community composition from 18 phylogenetically divergent sponge species in Vietnam. PeerJ.
2018;
6
:
e4970
.
View Article PubMed Google Scholar -
Brinkmann
M.C.,
Marker
A.,
Kurtböke
İ.D.,
An Overview on Marine Sponge-Symbiotic Bacteria as Unexhausted Sources for Natural Product Discovery. Diversity (Basel).
2017;
9
(4)
:
40
.
View Article Google Scholar -
Dat TTH, Hong TT, Dung TTK, Hoa NP, Cuong PV, Binh PT, Dat NT, Cuc NTK. Screening of antimicrobial producing bacteria associated with sponge and isolation of secondary metabolites from selected strain. Proceedings of National Conference on Biotechnology 2018; 2018 Oct 26; Hanoi, Vietnam. Hanoi, Vietnam: Publishing House for Science and Technology; 2018. p. 774-9.
.
-
Bibi
F.,
Faheem
M.,
Azhar
E.I.,
Yasir
M.,
Alvi
S.A.,
Kamal
M.A.,
Bacteria From Marine Sponges: A Source of New Drugs. Curr Drug Metab.
2017;
18
(1)
:
11-5
.
View Article PubMed Google Scholar -
Zhang
H.,
Zhao
Z.,
Wang
H.,
Cytotoxic Natural Products from Marine Sponge-Derived Microorganisms. Mar Drugs.
2017;
15
(3)
:
68
.
View Article PubMed Google Scholar -
Hong TT, Cuc NTK, Dat TTH, Cuong PV. Screening of anti-protease producing bacteria associated with marine sponges from Da Nang sea of Vietnam. Proceedings of National Conference on Biotechnology 2018; 2018 Oct 26; Hanoi, Vietnam. Hanoi, Vietnam: Publishing House for Science and Technology; 2018. p. 743-8.
.
-
Hong
T.T.,
Dat
T.T.H.,
Cuc
N.T.K.,
Cuong
P.V.,
Protease inhibitors from marine sponge and sponge-associated microorganisms. Vietnam Journal of Science and Technology..
2018;
56
(4)
:
405-23
.
View Article Google Scholar -
Pimentel-Elardo
S.M.,
Buback
V.,
Gulder
T.A.,
Bugni
T.S.,
Reppart
J.,
Bringmann
G.,
New tetromycin derivatives with anti-trypanosomal and protease inhibitory activities. Mar Drugs.
2011;
9
(10)
:
1682-97
.
View Article PubMed Google Scholar -
Wahyudi
A.T.,
Qatrunnada
Mubarik
N.R.,
Qatrunnada, Mubarik NR. Screening and Characterization of Protease Inhibitors from Marine Bacteria Associated with Sponge Jaspis sp. Hayati J Biosci.
2010;
17
(4)
:
173-8
.
View Article Google Scholar -
Zhang
T.,
Han
W.J.,
Liu
Z.P.,
Gene cloning and characterization of a novel esterase from activated sludge metagenome. Microb Cell Fact.
2009;
8
(1)
:
67
.
View Article PubMed Google Scholar -
Verma
D.,
Kawarabayasi
Y.,
Miyazaki
K.,
Satyanarayana
T.,
Cloning, expression and characteristics of a novel alkalistable and thermostable xylanase encoding gene (Mxyl) retrieved from compost-soil metagenome. PLoS One.
2013;
8
(1)
:
e52459
.
View Article PubMed Google Scholar -
Zhang
H.,
Li
F.,
Chen
H.,
Zhao
J.,
Yan
J.,
Jiang
P.,
Cloning, expression and characterization of a novel esterase from a South China Sea sediment metagenome. Chin J Oceanology Limnol.
2015;
33
(4)
:
819-27
.
View Article Google Scholar -
Singh
R.,
Dhawan
S.,
Singh
K.,
Kaur
J.,
Cloning, expression and characterization of a metagenome derived thermoactive/thermostable pectinase. Mol Biol Rep.
2012;
39
(8)
:
8353-61
.
View Article PubMed Google Scholar -
Maruthamuthu
M.,
van Elsas
J.D.,
Molecular cloning, expression, and characterization of four novel thermo-alkaliphilic enzymes retrieved from a metagenomic library. Biotechnol Biofuels.
2017;
10
(1)
:
142
.
View Article PubMed Google Scholar -
van Gent
D.,
Sharp
P.,
Morgan
K.,
Kalsheker
N.,
Serpins: structure, function and molecular evolution. Int J Biochem Cell Biol.
2003;
35
(11)
:
1536-47
.
View Article PubMed Google Scholar -
Jiang
C.J.,
Hao
Z.Y.,
Zeng
R.,
Shen
P.H.,
Li
J.F.,
Wu
B.,
Characterization of a novel serine protease inhibitor gene from a marine metagenome. Mar Drugs.
2011;
9
(9)
:
1487-501
.
View Article PubMed Google Scholar -
Chan
Y.S.,
Zhang
Y.,
Sze
S.C.,
Ng
T.B.,
A thermostable trypsin inhibitor with antiproliferative activity from small pinto beans. J Enzyme Inhib Med Chem.
2014;
29
(4)
:
485-90
.
View Article PubMed Google Scholar -
Shamsi
T.N.,
Parveen
R.,
Amir
M.,
Baig
M.A.,
Qureshi
M.I.,
Ali
S.,
Allium sativum Protease Inhibitor: A Novel Kunitz Trypsin Inhibitor from Garlic Is a New Comrade of the Serpin Family. PLoS One.
2016;
11
(11)
:
e0165572
.
View Article PubMed Google Scholar -
Mohan
M.,
Kozhithodi
S.,
Nayarisseri
A.,
Elyas
K.K.,
Screening, Purification and Characterization of Protease Inhibitor from Capsicum frutescens. Bioinformation.
2018;
14
(6)
:
285-93
.
View Article PubMed Google Scholar -
Shin
C.S.,
Hong
M.S.,
Bae
C.S.,
Lee
J.,
Enhanced production of human mini-proinsulin in fed-batch cultures at high cell density of Escherichia coli BL21(DE3)[pET-3aT2M2]. Biotechnol Prog.
1997;
13
(3)
:
249-57
.
View Article PubMed Google Scholar -
Piserchio
A.,
Ghose
R.,
Cowburn
D.,
Optimized bacterial expression and purification of the c-Src catalytic domain for solution NMR studies. J Biomol NMR.
2009;
44
(2)
:
87-93
.
View Article PubMed Google Scholar -
Tomala
M.,
Lavrentieva
A.,
Moretti
P.,
Rinas
U.,
Kasper
C.,
Stahl
F.,
Preparation of bioactive soluble human leukemia inhibitory factor from recombinant Escherichia coli using thioredoxin as fusion partner. Protein Expr Purif.
2010;
73
(1)
:
51-7
.
View Article PubMed Google Scholar -
Bacha
A.B.,
Jemel
I.,
Moubayed
N.M.S.,
Abdelmalek
I.B.,
Purification and characterization of a newly serine protease inhibitor from Rhamnus frangula with potential for use as therapeutic drug. 3 Biotech.
2017;
7
(2)
:
148
.
View Article PubMed Google Scholar -
Bijina
B.,
Chellappan
S.,
Basheer
S.M.,
Elyas
K.K.,
Bahkali
A.H.,
Chandrasekaran
M.,
Protease inhibitor from Moringa oleifera leaves: Isolation, purification, and characterization. Process Biochem.
2011;
46
(12)
:
2291-300
.
View Article Google Scholar -
Cruz
A.C.,
Massena
F.S.,
Migliolo
L.,
Macedo
L.L.,
Monteiro
N.K.,
Oliveira
A.S.,
Bioinsecticidal activity of a novel Kunitz trypsin inhibitor from Catanduva (Piptadenia moniliformis) seeds. Plant Physiol Biochem.
2013;
70
:
61-8
.
View Article PubMed Google Scholar -
Mello
G.C.,
Oliva
M.L.,
Sumikawa
J.T.,
Machado
O.L.,
Marangoni
S.,
Novello
J.C.,
Purification and characterization of a new trypsin inhibitor from Dimorphandra mollis seeds. J Protein Chem.
2001;
20
(8)
:
625-32
.
View Article PubMed Google Scholar -
Kidri\vc
M.,
Fabian
H.,
Brzin
J.,
Popovi\vc
T.,
Pain
R.H.,
Folding, stability, and secondary structure of a new dimeric cysteine proteinase inhibitor. Biochem Biophys Res Commun.
2002;
297
(4)
:
962-7
.
View Article PubMed Google Scholar -
Johnson
D.,
Travis
J.,
The oxidative inactivation of human alpha-1-proteinase inhibitor. Further evidence for methionine at the reactive center. J Biol Chem.
1979;
254
(10)
:
4022-6
.
PubMed Google Scholar
Comments

Article Details
Volume & Issue : Vol 7 No 2 (2020)
Page No.: 3633-3644
Published on: 2020-02-29
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
- HTML viewed - 8231 times
- Download PDF downloaded - 2065 times
- Supplementary 1 downloaded - 1569 times
- Supplementary 2 downloaded - 1292 times
- Supplementary 3 downloaded - 1541 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress