Abstract
Objective: Genetics is a known factor in the susceptibility to cardiovascular disease. In this study, polymorphisms of endothelial nitric oxide synthase (eNOS) (T876C and G894T) and hypoxiainducible factor 1 (HIF1) (rs10873142 and rs41508050) in coronary artery disease were compared with healthy subjects, and the relationship between these mutations and risk of coronary artery disease (CAD) was assessed.
Methods: Blood samples were collected from 138 patients with obstructive CAD and 115 healthy subjects. Polymerase chain reaction- restriction fragment length polymorphism (PCR-RFLP), Griess test, and enzyme-linked immunosorbent assay (ELISA) were used to measure serum nitric oxide (NO) and paraoxonase antioxidant. Independent t-test was used to examine the relationship between variables.
Results: There was no significant difference between genotype frequency in healthy subjects and patients for eNOS and HIF1 alpha polymorphisms. There was no significant difference between these polymorphisms and age, sex, hypertension and lipidemia. Furthermore, the difference in mean serum levels of NO in different genotypes of the G894T gene was not significant between the patient and control groups. However, the difference in mean serum NO levels in different genotypes of the T786C gene was significant between the patient and control groups such that minimum and maximum serum NO levels were observed in individuals bearing TT and TC genotypes, respectively. The difference in mean serum NO levels was higher in patients than in controls, and was statistically significant.
Conclusions: The results suggest that the T876C polymorphism could be associated with low serum NO level and implicated as a risk factor for developing CAD. Furthermore, our findings indicate that genetics may not be involved in susceptibility to CAD; however, further comprehensive studies are required.
Introduction
Cardiovascular diseases, especially coronary artery disease (CAD), are a major cause of death worldwide. CAD is a complex, multi-factorial and polygenic disease caused by atherosclerotic plaques in coronary artery walls. Generally, hypertension, hypercholesterolemia, smoking and diabetes are known as the main risk factors for cardiovascular disease. However, less than 10% of men with chronic heart disease have three or four risk factors concurrently and over 60% of cases have no or only one risk factor. On the other hand, some people with these risk factors lack cardiovascular complications. Therefore, genetics should also be considered as another important risk factor associated with cardiovascular complications. The role of genetics in cardiac disorders has long been unclear but recent studies indicate 45 hereditary factors predisposing to cardiovascular diseases 1.
Many studies suggest an equal contribution of genetic and environmental factors to the development of cardiovascular diseases and some researchers state that genetics plays the role of the trigger and environmental factors as shooting the gun 2. Hence, the results of investigations addressing real contribution of genetics are not conclusive, prompting us to study the role of genetics in the development of CAD in the almost genetically untouched Lorestan Province in the west of Iran. Given the fact that the rate of immigration to this province has been low and inhabited almost entirely by indigenous people, we hypothesized that our analysis would yield results different from studies conducted in Iran and worldwide. Indeed, our previous study in this province showed a significant difference in the prevalence of beta globin gene mutations in patients with thalassemia major in the Lorestan Province compared with that in most global investigations 3.
In the present research, we selected two genes of the antioxidant system which are effective on the cardiovascular system; the genes are hypoxia-inducible factor 1 alpha (HIF1α) and endothelial nitric oxide synthase (eNOS). The effects of important polymorphisms of these genes, such as C>T (rs10873142) and SNP: p.T4181 (rs41508050) in the HIF gene, as well as T786C and G894T in the eNOS gene, were studied in CAD patients.
Given the protective role of NO against important atherogenesis events, eNOS gene has gained considerable importance in the pathogenesis of coronary heart disease. Nitric oxide is a free radical and the most powerful vasodilator ever known. It plays an important role in the regulation of several physiological functions, including vascular activity, immunologic function, platelet aggregation inhibition, and limited low-density lipoprotein (LDL) oxidation 4. T786C polymorphism causes an approximately 50% reduction in eNOS gene promoter activity and substitution in exon 7 at codon 298 is associated with enzyme dysfunction 5. Several studies have reported the correlation between polymorphisms of oxidative genes, such as HIF1α and eNOS, with CAD 6 as well as the interaction of eNOS gene polymorphisms in CAD with serum NO levels 7.
HIF1 acts as a regulator of oxygen homeostasis. It plays an important role in atherosclerosis pathology through activation of several key genes involved in angiogenesis, energy metabolism, apoptosis/proliferation response, and vasomotor function 6. In fact, acute ischemic injury due to lack of oxygen homeostasis has been implicated in CAD pathogenesis. Disrupted balance in oxygen supply and demand is a function of reduced blood flow due to atherosclerotic plaque formation and inflammatory processes within the vascular endothelium 8.
In the study by Chen et al., HIF1α over-expression as well as correlation between HIF1α levels and severity of atherosclerosis have been reported 9. Interestingly, polymorphisms can regulate the expression, structure and stability of HIF1α mRNA or protein 10. Furthermore, eNOS is among the downstream target genes directly regulated by HIF1α 11, and is one of the three enzymatic isoforms which converts L-arginine to L-citrulline leading to NO synthesis 12.
The aim of the present study was to assess the impact of C>T (rs10873142) and p.T4181 SNP: (rs41508050) polymorphisms in the HIF gene as well as the T786C and G894T polymorphisms in the eNOS gene with regards to CAD susceptibility. Moreover, we aimed to study the relationship between different genotypes of T786C and G894T polymorphisms with concentrations of NO, and the difference in serum levels of NO and PON in the patient and control groups in Lorestan Province, Iran. The reason for choosing these mutations was that they had the highest association with heart disease in previous studies.
Methods
Subjects
This study was conducted in the Lorestan Province of Iran on 138 unrelated patients with CAD and 115 unrelated healthy subjects, from 2013 to 2015. Informed consent was obtained from patients and controls, and the study was approved by the Ethical Committee of Lorestan University of Medical Sciences. Patients supposedly with a history of CAD were referred to the angiography ward of the Cardiovascular Center at Shahid Madani Hospital in Khorramabad for further confirmation. Patients with established CAD were enrolled in the study. Healthy subjects were also enrolled after eligibility confirmation thorough examination.
The known risk factors for CAD are high blood pressure (≥ 90/140 mm Hg), diabetes mellitus (fasting glucose above 120 mg/dL or before treatment), family history of CAD, and smoking. These factors were assessed in both healthy subjects and patients. The case group included 73 men and 65 women with mean age of 61.53 ± 10.43 years, and the control group included 60 women and 55 men with mean age of 54.33 ± 13.41 years.
Phlebotomy
For genetic studies and measurement of serum biochemical parameters, venous blood samples were collected in the morning after overnight fasting and stored in EDTA-containing vials at 4°C. The vials were then centrifuged at 3500 rpm for 5 minutes and separated serum samples were kept at -80°C. The Geno Plus Genomic DNA Extraction Midiprep System (Viogene, Taiwan) was used for DNA extraction from blood samples, according to the manufacturer’s instructions.
Biochemical analysis
Serum NO level was determined by Griess method 13. Serum paraoxonase (PON1) level was measured using Human PON1 enzyme-linked immunosorbent assay (ELISA) Kit (Glory Science Co., Ltd, TX, USA) according to the kit protocol.
DNA analysis
We used polymerase chain reaction- restriction fragment length polymorphism (PCR-RFLP) technique to detect different SNP alleles and genotypes. The primer sequences, PCR conditions, and restriction enzymes (Fermentas) are summarized in Table 1 1415. The digestion products were then run on a 2% agarose gel and stained with safe stain.
| Variation | Primers | PCRconditions | PCRproduct size | Restrictionenzyme | RE digestion product size |
| HIF1α. C > T (rs10873142) | F-5' TTCTGTTCCTGGGTTATCTCAC-3' R-5'CCTTTAATGCAACAATGCCTAC-3' | 95°C/30 second 72°C/1 min | 864 bp | RsaI | WT-864 bpMut-707+157 bp |
| HIF1α.p.T4181 (rs41508050) | F-5' AATTTGGTGACATTTTGTTG-3' R-5' TGAGGGGAGCATTACATC-3' | 95°C/30 second 72°C/1 min | 492 bp | CaiI | WT- 436 + 56 bpMut- 492 bp |
| eNOS. T786C | F-5'TGGAGAGTGCTGGTGTACCCCA3'R-5'GCCTCCACCCCCACCCTGTC3' | 95°C/30 second 72°C/30 min | 180 bp | MspI | WT-140+40bpMut-90+50+40 bp |
| eNOS.G894T | F-5' AAGGCAGGAGACAGGATGGA3'R-5' CCCAGTCAATCCCTTTGGTGCTCA3' | 95°C/30 second 72°C/30 min | 248 bp | BanII | WT-163+85 bp Mut-248 bp |
Statistical analysis
SPSS version 18 was used for statistical analysis. The Hardy-Weinberg equilibrium for frequency of genotypes and characteristics of the subjects was reported with chi-square test. We used Fisher’s exact test and odds ratio (OD) with 95% confidence interval (CI) to calculate the association between CAD and the genetic polymorphisms. NO and PON1 concentrations were expressed as mean ±SD. Mean concentrations of PON1 and NO in serum, as well as NO concentrations of the different genotypes, were compared via analysis of variance (ANOVA) and t-tests. P<0.05 was considered statistically significant.
Results
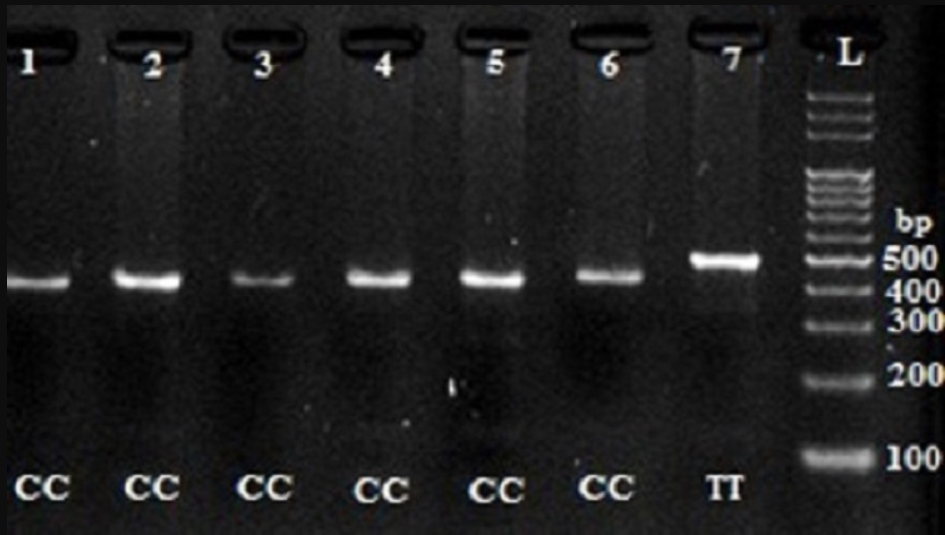
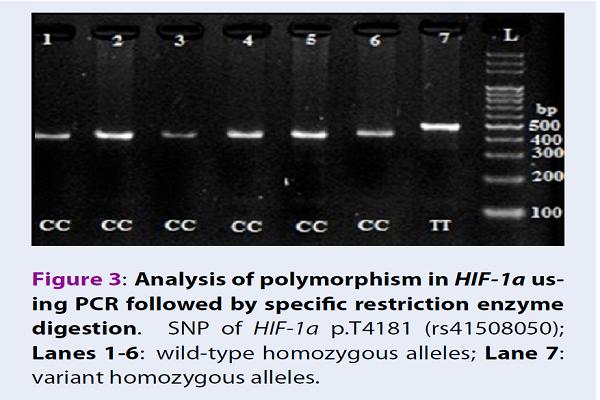
Digestion of wild-type and polymorphic DNA yields fragments which are shown in Figure 1Figure 2Figure 3Figure 4.
Characteristics of CAD patients and control subjects
The characteristics of patients and healthy subjects are demonstrated in Table 2. The two groups showed a significant difference in hypertension, hyperlipidemia and smoking (P <0.05).
| CAD, n | Controls, n | P-value | |
| Sex (Male/Female) | 65/73 | 55/60 | 0.90 |
| Age (Years) | 61.53 (±10.43) | 54.33 (±13.41) | 0.055 |
| Weight (Kg) | 71.20 (±16.41) | 70.00 (±10.59) | 0.62 |
| Hypertension (Yes/No)(> 140/90) | 81/57 | 33/82 | <0.001 |
| Hyperlipidemia (Yes/No)) | 57/81 | 22/93 | <0.001 |
| Diabetes mellitus (Yes/No)(Glucose of >120 mg/dL) | 19/119 | 8/107 | 0.081 |
| Smokers (Yes/No) | 36/102 | 14/100 | 0.006 |
| CAD Heredity (Yes/No) | 37/101 | 33/82 | 0.73 |
Distribution of genotypes and alleles of HIF1α and eNOS polymorphisms in patients vs. controls
Distribution of genotype polymorphisms did not indicate deviation from the Hardy-Weinberg equilibrium (P>0.05). The frequency distribution of genotypes for the patient and control groups for 4 polymorphisms was compared in Table 3.
| Genotype | CAD n (%) | Controls n (%) | OR | CI 95% | P-value | |
| C > T (rs10873142) | TT (wt/wt) | 58 (43.9) | 45 (40.2) | Ref | ||
| TC (wt/mut) | 62 (47) | 58 (51.8) | 0.82 | (0.48-1.40) | 0.48 | |
| CC (mut/mut) | 12 (9.1) | 9 (8) | 1.03 | (0.40-2.66) | 1 | |
| Alleles | T | 178 (67.4) | 148 (66.1) | Ref | ||
| C | 86 (32.6) | 76 (33.9) | 0.94 | (0.64-1.37) | 0.75 | |
| p.T 418 1 (rs41508050) | CC (wt/wt) | 136 (98.55) | 113 (98.25) | Ref | ||
| CT (wt/mut) | 0 | . | - | - | - | |
| TT (mut/mut) | 2 (1.44) | 2 (1.74) | 0.83 | (0.11-6.00) | 1 | |
| Alleles | C | 272 (98.55) | 226 (98.7) | Ref | ||
| T | 4 (1.45) | 4 (1.3) | 1.11 | (0.24-5.02) | 1 | |
| T786C | TT (wt/wt) | 74 (54) | 57 (50) | Ref | ||
| TC (wt/mut) | 55 (40.1) | 47 (41.2) | 0.90 | (0.53-1.51) | 0.69 | |
| CC (mut/mut) | 8 (5.8) | 10 (8.8) | 0.61 | (0.22-1.66) | 0.33 | |
| Alleles | T | 203 (74.08) | 161 (70.61) | Ref | ||
| C | 71 (25.91) | 67 (29.38) | 0.84 | (0.56-1.24) | 0.38 | |
| G894T | GG (wt/wt) | 74 (59.2) | 57 (52.3) | Ref | ||
| GT (wt/mut) | 40 (32) | 39 (35.8) | 0.79 | (0.45-1.38) | 0.40 | |
| TT (mut/mut) | 11 (8.8) | 13 (11.9) | 0.65 | (0.27-1.56) | 0.33 | |
| Alleles | G | 188 (75.20) | 153 (70.18) | Ref | ||
| T | 62 (24.80) | 65 (29.82) | 0.77 | (0.51-1.16) | 0.22 |
No significant difference was observed between the two groups for the 4 polymorphisms in terms of genotype and allele frequencies (all were P>0.05). Frequency comparison of C>T (rs10873142) polymorphism genotype of HIF1α gene in the two groups showed that the frequency of TC genotype was 47% and 51.8% in patients and controls, respectively. The CC genotype had the lowest frequency in patients (8%) and controls (9.1%), but the difference in frequency distribution of C>T (rs10873142) genotype in the two groups was not statistically significant.
The frequency of CC, TC and TT genotypes in p.T4181 (rs 41508050) polymorphism was 136 (98.55%) and 0 (0%), 2 (1.44%) and 113 (98.25%), and 1 (0.87%) and 1 (0.87%) in patient and control groups, respectively, for the genotypes. The difference in frequency distribution of p.T418I (rs41508050) genotypes in the two groups (patient and control) was not statistically significant.
The frequency of TT, TC and CC genotypes in the T786C polymorphism in patient group was 74 (54%) and 55 (40.1%), 8 (5.8%); whereas in control group it was 57(50%), 47 (41.2%) and 10 (8.80%), respectively.
The comparison of frequency of T786C genotypes between patient and control groups revealed that the TT genotype had the highest frequency (54% and 50%, respectively), while CC genotype had the lowest frequency in patient and control groups (54% and 50%, respectively). However, the difference in frequency distribution of T786C genotypes in the two groups was not statistically significant (P>0.05).
The frequency of GG, GT and TT genotypes in G894 polymorphism in patient group was 74 (59.2%), 40 (32%),11 (8.8%); while it was 57(52.3%), 39 (35.8%) and 13 (11.9%) control group, respectively.
Frequency comparison of G894 genotypes in patient and control groups showed that GG genotype had the highest frequency (59.2% and 52.3%, respectively) and TT genotype had the lowest frequency in patient and control groups (8.8% and 11.9%, respectively), but the difference in frequency distribution of G894T genotypes between the two groups was not statistically significant (Table 3).
Effects of T786C and G894T polymorphisms of eNOS on serum NO concentration
The difference in mean NO levels between healthy subjects and patients with different genotypes of T786C polymorphism was statistically significant (P=0.001), such that the highest and lowest NO levels were observed in individuals with TT and TC genotype, respectively (Table 4). Moreover, the difference in mean serum NO levels for the genotypes of G894T polymorphism in patients and controls was not statistically significant.
| T786C | TT | TC | CC | p-value |
| Cases. Nitric oxide (NO)(×10 2 µmol/L) | 0.16 ± 1.55 | 0.21 ± 1.38 | 0.12 ± 1.49 | 0.001 |
| Controls. Nitric oxide (NO) (×10 2 µmol/L) | 0.11 ± 1.47 | 0.11 0± 1.34 | 0.09 ± 1.41 | 0.001 |
| G894T | GG | GT | TT | p-value |
| Case Nitric oxide (NO) (×10 2 µmol/L) | 0.18 ± 1.49 | 0.19 ± 1.48 | 0.17 ± 1.39 | 0.28 |
| Control Nitric oxide(NO) (×10 2 µmol/L) | 0.13 ± 1.40 | 0.13 ± 1.43 | 0.10 ± 1.40 | 0.48 |
Mean serum NO and PON1 concentration in case and control groups
The mean difference in serum NO levels between patients (1.48 ± 0.19 ×102 µmol/L) and controls (1.41 ± 0.12×10 2 µmol/L) was statistically significant (P=0.002); NO level was higher in the patient group than control. Mean serum concentration of PON1 enzyme was lower in patients (17.89 ± 13.97 nmol/L) than in the controls (21.23 ± 13.83 nmol/L), but the difference was not statistically significant (P=0.272).
Discussion
CAD is a multi-factorial disease of which several genetic and environmental factors, as well as ethnic susceptibility, are involved in its development 12. Understanding the genetic factors involved in susceptibility to a disease results in better treatment options for the disease 16. In this study, no significant association was found between C>T (rs10873142) and p.T4181 (rs41508050) polymorphisms of the HIF-1α gene with CAD. Different results have been reported in the study of association between HIF1α polymorphisms and cardiovascular diseases. There have been correlations reported for rs10873142, rs41508050 and rs11549465 polymorphisms with cardiovascular diseases, such as ischemic heart disease, CAD with stable exertional angina, and acute myocardial infraction 17. However, no association was found for rs2057482, rs11549467 and rs11549465 polymorphisms with these diseases 18.
The findings of other studies on the relationship between eNOS gene polymorphisms and CAD are not consistent. For instance, in 1998, Hibi et al. pointed out, for the first time, the relationship between NO synthase gene polymorphism and myocardial infarction 19. Results from a study by Nakayama et al. suggests that the CC variant in T786C polymorphism reduces eNOs synthesis and predisposes the patients to coronary spasms 20. A study on Caucasian population showed that the C allele is associated with increased risk of multivessel CAD 21. Tangurek et al. also found a significant difference in the distribution of T786C genotypes between patients and healthy subjects 22. However; similar to results of Granath et al. 23 and Ciftci et al. 24, we found no significant difference in the frequency of T786C genotypes between CAD patients and those in the control group.
Furthermore, the frequencies of the T786C polymorphism genotypes in healthy subjects and patient groups were similar, but in general, a slight increase in the frequency of wild TT genotype was observed in patients. In a study in Iran, a significant increase in T mutant frequency was reported in patients compared to controls 25.
In our study of the G894T polymorphism, genotypes frequency was similar in healthy subjects and patients but, in general, a slight increase was seen in the frequency of wild GG genotype in patients. In general, few studies have been conducted on the relationship between eNOS gene polymorphisms and NO serum levels. In a study in UK evaluating the effect of G894T, T786C, VNTR intron 4 and A922G polymorphisms on NO production, it was concluded that none of these polymorphisms affect the plasma NO production 26. Similar to our study, in a study in Turkey, a correlation between CAD and T mutant allele in the G894T polymorphism, as well as serum levels of NO, was not confirmed 27. Unlike our study, Yoon et al. reported increased serum levels of NO in control subjects bearing G894T mutation 28.
In some studies, it has been reported that the T786C polymorphism does not affect plasma NO concentrations. Meanwhile in other studies, the association between reduced NO concentration in blood circulation and the presence of eNO-specific haplotypes bearing C (T786C) and G (G894T) alleles have been mentioned.
In our study, the difference in mean NO levels in the different genotypes of T786C gene was statistically significant (P<0.05) in both healthy subjects and patients, in such a way that maximum and minimum NO level was observed in individuals with TT and TC genotypes, respectively. In general, NO serum level was lower in individuals with mutant C alleles (heterozygotes and mutant homozygotes) than those with wild homozygous alleles. The results of correlation between serum levels of NO and CAD are different (similar to results of gene distribution and frequency of polymorphisms). In this study, we saw a significant increase in mean serum levels of NO in CAD patients (1.48 × 102 ± 0.19 μmol/L) compared to healthy controls (1.41 × 102 ± 0.12 μmol/L).
NO plays an important role in atherogenesis, and atherosclerosis has been associated with decreased endothelial NO production. In these pathological conditions, the expression of eNOS may be increased and result in increased NO levels. These findings suggest that an increase in serum levels of NO in CAD patients can play a compensatory role for the resumption of endothelial vasomotor function 2425262728.
Increased oxidative stress or reduced antioxidant defense mechanisms may be involved in the onset and progression of atherosclerosis. In some studies, the relationship between reduced levels of PON1 with CAD has been mentioned, but Rahmani and Azizi (in separate studies on Iranian population) have reported no significant differences in PON1 activity in patients and controls 293031. In our study, although serum levels of PON1 in the patient group were lower than the control group, no significant difference was found in serum PON1 levels between the two groups.
Moreover, we found no correlation between the frequency of genotypes and mutant alleles of T786C and G894T polymorphisms with CAD. Likewise, no correlation was found between G894T genotypes and serum NO levels in both groups. However, the difference of mean serum NO levels in T786C polymorphism in different genotypes was significant in both control and patient groups. In general, serum NO levels in individuals with mutant C allele (heterozygotes and homozygous mutants) were lower than individuals with wild-type homozygous allele.
Conclusions
Our results show that serum NO levels are relatively low in mutant C allele in T786C polymorphisms. As previously mentioned, inconclusive results suggest a correlation between NO gene polymorphisms, as well as NO serum levels, with cardiovascular disease. One possible explanation for these conflicting results could be that such studies usually concentrate on only one polymorphism in certain clinical conditions and the average impact may not be identified. Ethnic differences and the size of population under study can also impact inconsistent results from different studies.
To reinforce our understanding of the contribution of gene polymorphisms explored in this study and also to achieve any potential clinical relevance in terms of prognosis or therapeutic intervention for CAD patients, similar studies on these genes in different and larger populations are necessary.
Competing Interests
The authors declare no conflict of interest.
Authors' Contributions
Ali Asghar Kiani: conceived of the presented idea and developed the theory and performed the computations;
Vahide Heydari Nazarabad: carried out the experiment;
Kolsum Ahmadi: carried out the experimen;
Khatereh Anbari: verified the analytical methods;
Babak Baharvand Ahmadi: supervised the findings of this work.
All authors discussed the results and contributed to the final manuscript.
Acknowledgments
The project was sponsored by Deputy Office for Research of the Lorestan University of Medical Sciences.
Data Availability
The data used to support the findings of this study are included within the article.
References
-
Anderson
J. L.,
Carlquist
J. F.,
Horne
B. D.,
Hopkins
P. N..
Progress in unraveling the genetics of coronary artery disease and myocardial infarction. Current Atherosclerosis Reports.
2007;
9
:
179-86
.
-
Kessler
T.,
Erdmann
J.,
Schunkert
H..
Genetics of coronary artery disease and myocardial infarction—2013. Current Cardiology Reports.
2013;
15
:
368
.
-
Kiani
A. A.,
Mortazavi
Y.,
Zeinali
S.,
Shirkhani
Y..
The molecular analysis of beta-thalassemia mutations in Lorestan Province, Iran. Hemoglobin.
2007;
31
:
343-9
.
-
Moncada
S.,
Palmer
R. M.,
Higgs
E. A..
Nitric oxide: physiology, pathophysiology, and pharmacology. Pharmacological Reviews.
1991;
43
:
109-42
.
-
Tesauro
M.,
Thompson
W. C.,
Rogliani
P.,
Qi
L.,
Chaudhary
P. P.,
Moss
J..
Intracellular processing of endothelial nitric oxide synthase isoforms associated with differences in severity of cardiopulmonary diseases: cleavage of proteins with aspartate vs. glutamate at position 298. Proceedings of the National Academy of Sciences of the United States of America.
2000;
97
:
2832-5
.
-
Colombo
M. G.,
Andreassi
M. G.,
Paradossi
U.,
Botto
N.,
Manfredi
S.,
Masetti
S..
Evidence for association of a common variant of the endothelial nitric oxide synthase gene (Glu298—>Asp polymorphism) to the presence, extent, and severity of coronary artery disease. Heart (British Cardiac Society).
2002;
87
:
525-8
.
-
Tsukada
T.,
Yokoyama
K.,
Arai
T.,
Takemoto
F.,
Hara
S.,
Yamada
A..
Evidence of association of the ecNOS gene polymorphism with plasma NO metabolite levels in humans. Biochemical and Biophysical Research Communications.
1998;
245
:
190-3
.
-
López-Reyes
A.,
Rodríguez-Pérez
J. M.,
Fernández-Torres
J.,
Martínez-Rodríguez
N.,
Pérez-Hernández
N.,
Fuentes-Gómez
A. J..
The HIF1A rs2057482 polymorphism is associated with risk of developing premature coronary artery disease and with some metabolic and cardiovascular risk factors. The Genetics of Atherosclerotic Disease (GEA) Mexican Study. Experimental and Molecular Pathology.
2014;
96
:
405-10
.
-
Chen
S. M.,
Li
Y. G.,
Wang
D. M.,
Zhang
G. H.,
Tan
C. J..
Expression of heme oxygenase-1, hypoxia inducible factor-1alpha, and ubiquitin in peripheral inflammatory cells from patients with coronary heart disease. Clinical Chemistry and Laboratory Medicine.
2009;
47
:
327-33
.
-
Prior
S. J.,
Hagberg
J. M.,
Phares
D. A.,
Brown
M. D.,
Fairfull
L.,
Ferrell
R. E..
Sequence variation in hypoxia-inducible factor 1alpha (HIF1A): association with maximal oxygen consumption. Physiological Genomics.
2003;
15
:
20-6
.
-
Coulet
F.,
Nadaud
S.,
Agrapart
M.,
Soubrier
F..
Identification of hypoxia-response element in the human endothelial nitric-oxide synthase gene promoter. The Journal of Biological Chemistry.
2003;
278
:
46230-40
.
-
Mayer
B.,
Hemmens
B..
Biosynthesis and action of nitric oxide in mammalian cells. Trends in Biochemical Sciences.
1997;
22
:
477-81
.
-
Miranda
K. M.,
Espey
M. G.,
Wink
D. A..
A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide.
2001;
5
:
62-71
.
-
Konac
Ece,
Dogan
Irem,
Onen
Hacer Ilke,
Yurdakul
Ahmet Selim,
Ozturk
Can,
Varol
Ayhan,
Ekmecki
Abdullah.
Genetic Variations in the Hypoxia-Inducible Factor-1$α$Gene and Lung Cancer. Experimental Biology and Medicine.
2009;
234
(9)
:
1109-1116
.
-
Marroni
A. S.,
Metzger
I. F.,
Souza-Costa
D. C.,
Nagassaki
S.,
Sandrim
V. C.,
Correa
R. X..
Consistent interethnic differences in the distribution of clinically relevant endothelial nitric oxide synthase genetic polymorphisms. Nitric Oxide.
2005;
12
:
177-82
.
-
Exner
M.,
Minar
E.,
Wagner
O.,
Schillinger
M..
The role of heme oxygenase-1 promoter polymorphisms in human disease. Free Radical Biology & Medicine.
2004;
37
:
1097-104
.
-
Zheng
Z. L.,
Hwang
Y. H.,
Kim
S. K.,
Kim
S.,
Son
M. J.,
Ro
H..
Genetic polymorphisms of hypoxia-inducible factor-1 alpha and cardiovascular disease in hemodialysis patients. Nephron. Clinical Practice.
2009;
113
:
c104-11
.
-
Alidoosti
M.,
Ghaedi
M.,
Soleimani
A.,
Bakhtiyari
S.,
Rezvanfard
M.,
Golkhu
S..
Study on the role of environmental parameters and HIF-1A gene polymorphism in coronary collateral formation among patients with ischemic heart disease. Clinical Biochemistry.
2011;
44
:
1421-4
.
-
Hibi
K.,
Ishigami
T.,
Tamura
K.,
Mizushima
S.,
Nyui
N.,
Fujita
T..
Endothelial nitric oxide synthase gene polymorphism and acute myocardial infarction. Hypertension.
1998;
32
:
521-6
.
-
Nakayama
M.,
Yasue
H.,
Yoshimura
M.,
Shimasaki
Y.,
Kugiyama
K.,
Ogawa
H..
T-786—>C mutation in the 5′-flanking region of the endothelial nitric oxide synthase gene is associated with coronary spasm. Circulation.
1999;
99
:
2864-70
.
-
Rossi
G. P.,
Cesari
M.,
Zanchetta
M.,
Colonna
S.,
Maiolino
G.,
Pedon
L..
The T-786C endothelial nitric oxide synthase genotype is a novel risk factor for coronary artery disease in Caucasian patients of the GENICA study. Journal of the American College of Cardiology.
2003;
41
:
930-7
.
-
Tangurek
B.,
Ozer
N.,
Sayar
N.,
Terzi
S.,
Yilmaz
H.,
Dayi
S. U..
The relationship between endothelial nitric oxide synthase gene polymorphism (T-786 C) and coronary artery disease in the Turkish population. Heart and Vessels.
2006;
21
:
285-90
.
-
Granath
B.,
Taylor
R. R.,
van Bockxmeer
F. M.,
Mamotte
C. D..
Lack of evidence for association between endothelial nitric oxide synthase gene polymorphisms and coronary artery disease in the Australian Caucasian population. Journal of Cardiovascular Risk.
2001;
8
:
235-41
.
-
Ciftçi
C.,
Melil
S.,
Cebi
Y.,
Ersöz
M.,
Cağatay
P.,
Kiliçgedik
M..
Association of endothelial nitric oxide synthase promoter region (T-786C) gene polymorphism with acute coronary syndrome and coronary heart disease. Lipids in Health and Disease.
2008;
7
:
5
.
-
Mehrtashfar
Shokoufeh,
Khatir
Aria Esmaili,
Ebrahimi
Ahmad.
Association of enos 894 g> t gene polymorphism and risk of coronary artery disease in iranian population. 2014
.
-
Jeerooburkhan
N.,
Jones
L. C.,
Bujac
S.,
Cooper
J. A.,
Miller
G. J.,
Vallance
P..
Genetic and environmental determinants of plasma nitrogen oxides and risk of ischemic heart disease. Hypertension.
2001;
38
:
1054-61
.
-
Afrasyap
L.,
Ozturk
G..
NO level and endothelial NO synthase gene polymorphism (Glu298Asp) in the patients with coronary artery disease from the Turkish population. Acta Biochimica et Biophysica Sinica.
2004;
36
:
661-6
.
-
Yoon
Y.,
Song
J.,
Hong
S. H.,
Kim
J. Q..
Plasma nitric oxide concentrations and nitric oxide synthase gene polymorphisms in coronary artery disease. Clinical Chemistry.
2000;
46
:
1626-30
.
-
Rahmani
M.,
Raiszadeh
F.,
Allahverdian
S.,
Kiaii
S.,
Navab
M.,
Azizi
F..
Coronary artery disease is associated with the ratio of apolipoprotein A-I/B and serum concentration of apolipoprotein B, but not with paraoxonase enzyme activity in Iranian subjects. Atherosclerosis.
2002;
162
:
381-9
.
-
Azizi
F.,
Rahmani
M.,
Raiszadeh
F.,
Solati
M.,
Navab
M..
Association of lipids, lipoproteins, apolipoproteins and paraoxonase enzyme activity with premature coronary artery disease. Coronary Artery Disease.
2002;
13
:
9-16
.
-
Soltanpour
Mohammad Soleyman,
Soheili
Zahra,
Pourfathollah
Ali Akbar,
Samiei
Shahram,
Meshkani
Reza,
Kiani
Ali Asghar,
Majid
Safa,
Ataei
Zahra,
Kavari
Mahnaz.
The A1298C mutation in methylenetetrahydrofolate reductase gene and its association with idiopathic venous thrombosis in an Iranian population. Laboratory Medicine.
2011;
42
(4)
:
213-216
.
Comments
Downloads
Article Details
Volume & Issue : Vol 5 No 9 (2018)
Page No.: 2688-2696
Published on: 2018-09-29
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 6135 times
- Download PDF downloaded - 1891 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress