Abstract
Background: Ion channels play a crucial role in Glomerular filter damage that contributes to albuminuria. Transient receptor potential channel 5 (TRPC5) gene mediating such damage, demand for its target specific inhibition by RNA interference mechanism. Designing and selecting potential siRNA for TRPC5 gene silencing by computational analysis.
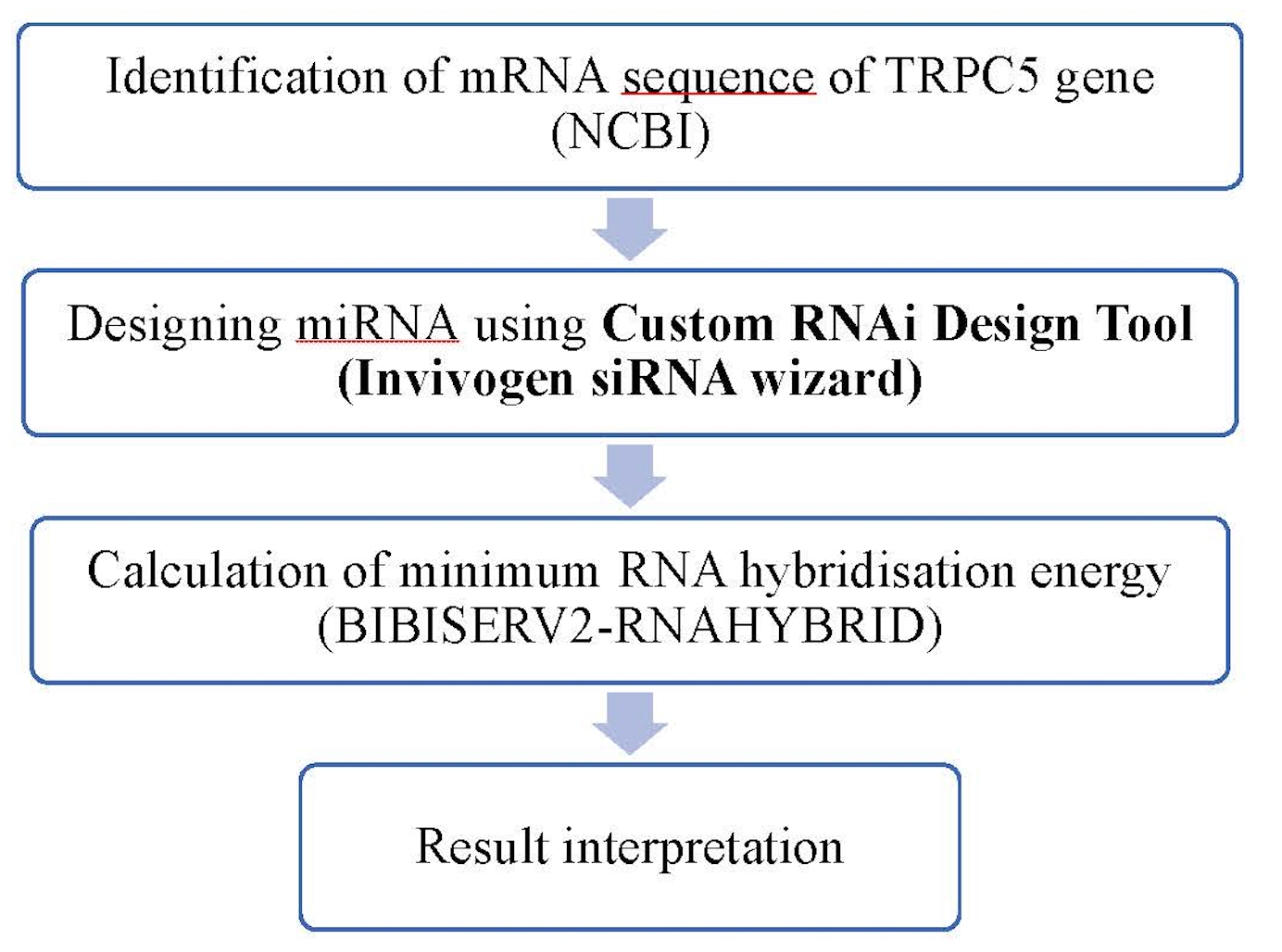
Materials & Methods: The mRNA sequence was retrieved from NCBI (National Center for Biotechnology Information). siRNA sequences were designed specifically from target genes using InvivoGen siRNA wizard software. Thermodynamic RNA-RNA interactions were used to evaluate the gene silencing efficiency by minimum free energy of hybridization; the hybridization structures were also obtained using BIBISERV2-RNAHybrid.
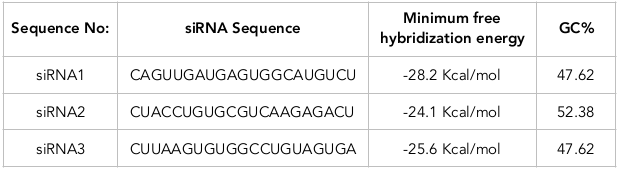
Results: The minimum free energy of hybridization of the three designed siRNAs (siRNA1, siRNA2 and siRNA3) were as follows: -28.2 kcal/mol, -24.1 kcal/mol, and-25.6 kcal/mol. Their corresponding GC content were 47.62%, 52.38% and 47.62%, respectively. Thus, siRNA1 had the least minimum free energy of hybridization (i.e. -28.2 kcal/mol) with low GC content (47.62%), and high linearity with minimal h-b index and loop structure.
Conclusion: RNAi therapy can provide a new platform for efficient and targeted therapeutics. Further in vivo investigations are necessary to further validate their efficacy.
Introduction
Albuminuria is a major cause of morbidity and mortality in patients with chronic kidney diseases (CKD). It is a well-known condition that contributes to cardiovascular disease, poor renal outcome, diabetes, hypertension and kidney failure Mogensen, 1984Berrut et al., 1997Keane et al., 2003Anavekar et al., 2004Gerstein et al., 2001. One of the key causes of albuminuria is malfunction of the glomerular filter which is crucial in sorting and preventing essential molecules from spilling into urine. Currently, there are no targeted therapies for the efficient protection of filter barrier function. The human kidney filter handles 180L of plasma each day. Therefore, an intact kidney filter is essential for retention of essential proteins in blood and removal of waste from the body Haraldsson, 2008Farquhar, 2006.
Ion channels play a predominant role in regulation of kidney filter function. Transient receptor potential (TRP) channels are greatly conserved nonselective cationic channels first documented in Drosophila, that has 6 members forming either homomeric or heteromeric tetramer Montell, 2005Ramsey et al., 2006. Since these channels are greatly expressed in brain and kidney, their loss has the ability to cause fear response in mice Riccio et al., 2009. Transient receptor potential channel 5 (TRPC5) functions as a cellular sensor of redox changes with inference for inflammation Xu et al., 2008. Calcium (Ca2+) influx through homomeric TRPC5 channels alter the Rho family GTPase Rac1 to interrupt the integrity of the actin cytoskeleton in mesenchymal cells, such as fibroblasts and podocytes, at the cellular level resulting in filtration barrier damage. Therefore, Rac signaling can direct albuminuria in mice Tian et al., 2010Faul et al., 2008Ma et al., 2010, Yanagida-Asanuma et al., 2007Togawa et al., 1999. TRPC5 intercedes damage to the filtration barrier, contributing to albuminuria which serves as an indicator for cardiovascular, metabolic and chronic kidney diseases. Hence, inhibition of the TRPC5 gene is a specific approach to protect glomerular filter damage and albuminuria Thomas schaldecker et al., 2013.
RNA interference (RNAi) is an evolutionary conserved mechanism of gene regulation process that needs double stranded RNA processed into small interfering RNA (siRNA) and micro RNA (miRNA) to suppress the expression of target genes (i.e. mRNA) in a sequence specific manner Tuschl and Borkhardt, 2002, Ma et al., 2007. An siRNA is a double stranded RNA molecule typically having 19-21 nucleotide base pairs, that mediate its binding with target mRNA and promotes its degradation. The degradation prevents the translation of the mRNA, thus repressing the activity of the targeted mRNA Elbashir et al., 2001Zamore et al., 2000. siRNA can be chemically synthesized and delivered into the cells by direct transfection, using nanoparticles or plasmid/viral vectors (Sayda et al., 2001; Brummelkamp et al., 2002). Once the siRNA enters the cells, it gets cleaved by dicer (RNase III-like enzyme) and gets integrated into RNA-induced silencing complex (RISC) where the sense (passenger) stranded is degraded within RISC, while the anti-sense strands proceeds for post-transcriptional gene silencing process Jackson et al., 2010. The degradation process is an extremely complicated one that consists of multiple steps including the initial binding of siRNA to RISC, followed by its activation which leads to mRNA recognition and degradation by using different exo- and endo- nucleases Bernstein et al., 2001Hammond et al., 2000. The aim of this study is to inhibit TRPC5 gene in a target specific approach by designing effective siRNA molecules in context of RNAi mechanism.
Materials - Methods
Retrieval of TRPC5 gene (mRNA) sequence
The mRNA sequence of the TRPC5 gene was retrieved from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/). The Accession number of TRPC5 mRNA coding sequence was NM_012471.2. The sequence was retrieved in FASTA format and used for designing sequence specific siRNA molecules.
Design of Potential siRNA molecules
Target recognition was designed for the target TRPC5 mRNA using an online bioinformatic tool called InvivoGen siRNA wizard (http://www.invivogen.com/sirnawizard/design.php). This tool utilizes definite parameters such as siRNA motif size (21 nucleotides) and mRNA database (for human). siRNA sequences containing a palindrome are excluded to avoid unwanted hairpin structures. siRNA duplex with low GC content ranging from 30-55% are selected. The tool employs blast search to reduce off-target similarity.
Multiple sequence alignment of designed siRNA molecules
To analyze the sequence similarity and evolutionary relationship between designed siRNA molecules, multiple sequence alignment was conducted using ClustalW tool (http://www.ebi.ac.uk/Tools/msa/; http://www.clustal.org/) according to standard parameters. Phylogenetic tree was constructed using clustal tree format, kimura’s distance correction and UPGMA (unweighted pair group method with arithmetic mean) method. Percentage identity matrix was also calculated.
Calculation of GC content
The GC content of the designed siRNA molecules was calculated using online GC calculator (http://www.endmemo.com/bio/gc.php). Any siRNA duplex with low GC content ranging from 40-55% was selected for further screening.
The GC content was calculated using the following formula:
(Number of G nucleotide + Number of C nucleotide)/Total number of nucleotide X 100 = GC content %
Screening of designed siRNAs
Each designed siRNA was screened based on thermodynamic evaluation of RNA-RNA interaction using the online bioinformatic software BIBISERV2-RNAHYBRID Rehmsmeier et al., 2004. Thermodynamic interaction was studied between predicted siRNA (guide strand) and target gene. A dynamic programming algorithm in the software was used to calculate the hybridization energy and base pairing form of two RNA sequences. The software also provides the hybridization structure for further evaluation. A flow chart representing the complete methodology used for screening of effective siRNA molecules against TRPC5 mRNA is illustrated in Figure 1 .
Results
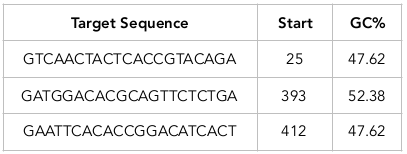
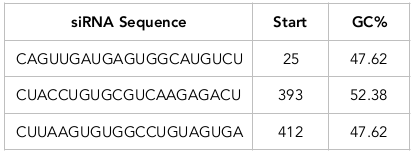
According to siRNA tool results, three siRNAs have been designed from target mRNA sequences ( Table 1 ; Table 2 ). The tool predicts the target sites from mRNA sequences for siRNA design. siRNA1 was designed from target sequence GTCAACTACTCACCGTACAGA at a site starting at the 25th position on the mRNA. The guide strand of the siRNA1 has a sequence complementary to the target site sequence (CAGUUGAUGAGUGGCAUGUCU). Similarly, siRNA 2 and siRNA 3 have target sequences GATGGACACGCAGTTCTCTGA and GAATTCACACCGGACATCACT, respectively, at sites starting at the 393th and 412th position on mRNA, respectively. The guide strands of siRNA 2 and siRNA 3 have the sequence CUACCUGUGCGUCAAGAGACU and CUUAAGUGUGGCCUGUAGUGA. Multiple sequence alignments showed the presence of a conserved region among the functional siRNAs.
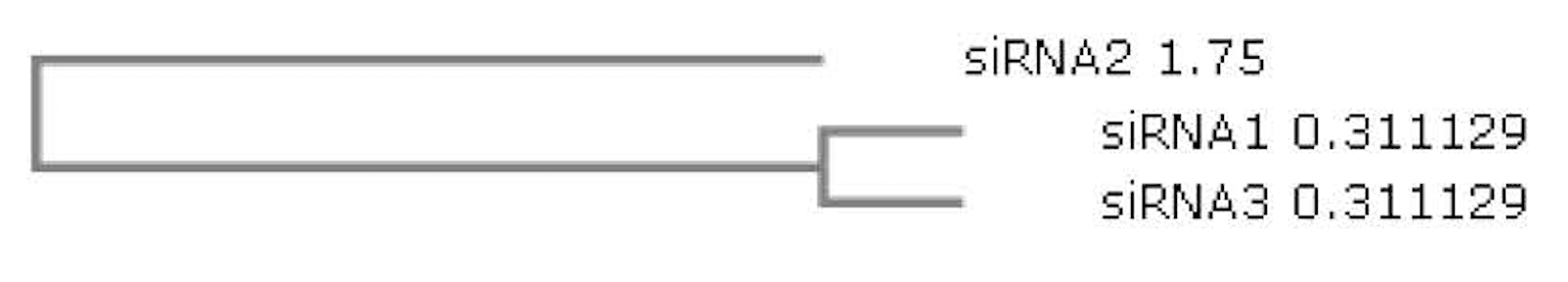
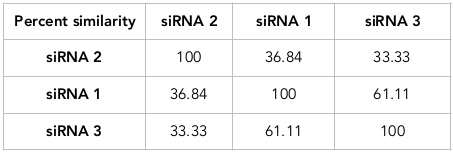
Figure 2 represent the phylogenetic relationship between the designed siRNAs. The branch length of the siRNA1, siRNA2 and siRNA3 are 0.311129, 1.75 and 0.311129, respectively. Their sequence similarity was evaluated using percent identity matrix ( Table 4 ). siRNA 2 shows 36.84% and 33.33% similarity to siRNA 1 and siRNA 3, respectively. siRNA 1 is similar to siRNA 2 and siRNA 3 with 36.84% and 61.11% similarity, respectively. Additionally, siRNA 3 is similar to siRNA 1 and siRNA 2 with 33.33% and 61.11% similarity, respectively.
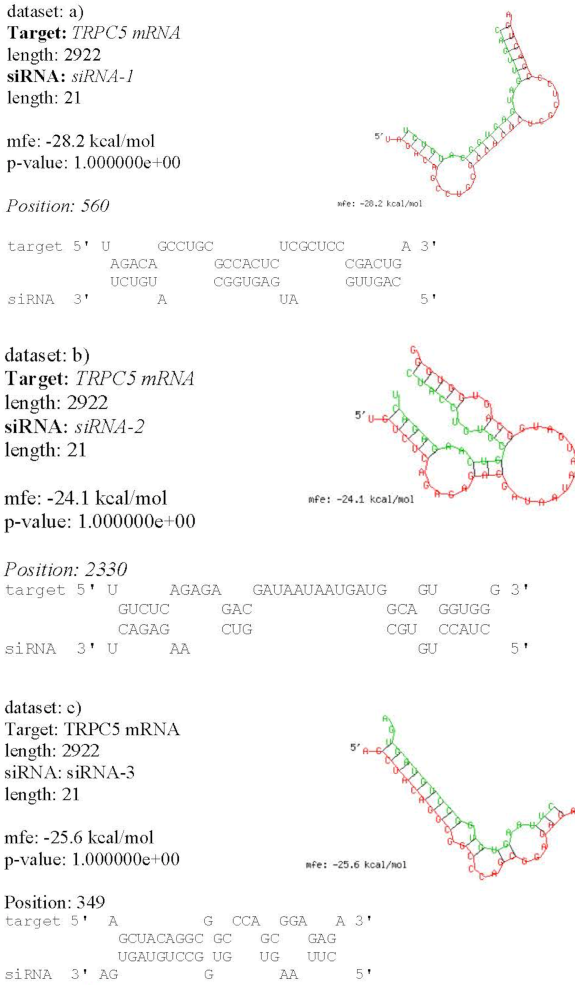
The gene silencing efficacy of siRNA was evaluated based on the thermodynamic interaction between the target TRPC5 mRNA and the designed guide strands of siRNA. The minimum free energy of hybridization of the three designed siRNAs (e.g. siRNA1, siRNA2 and siRNA3) were as follows: -28.2 kcal/mol, -24.1 kcal/mol and -25.6 kcal/mol. Their corresponding GC content for each was 47.62%, 52.38% and 47.62%, respectively ( Table 3 ). The hybridization structure was observed to evaluate further parameters such as linearity ( Figure 3 ). From all these, sequence #1 (i.e. siRNA1) is a more suitable and accessible one since it has the least minimum free hybridization energy (-28.2 kcal/mol). Indeed, it is predicted to be the most efficient method towards TRPC5 gene silencing.
Discussion
This study was conducted with retrieval of TRPC5 mRNA sequence from NCBI. siRNA wizard tool has been used to identify potential, target specific siRNA molecules (from target mRNAs) that significantly reduces off-target silencing. The tool utilizes selection criteria for designing potential siRNA such as thermodynamics, GC content analysis, BLAST search, secondary structure avoidance, termination signal, and immunostimulatory motif exclusion (http://www.invivogen.com/sirna-wizard)
All the 3 siRNA sequences were designed specifically to have low off target similarity which can be suitable for effective post-transcriptional gene silencing process. Multiple sequence alignment (via ClustalW tool) was used to check the sequence similarity across the siRNAs. A phylogenetic tree was constructed to study the evolutionary relationship and the consensus sequence between the designed functional low off target siRNA molecules. The analysis revealed the percentage similarity between the sequences with branch length shown in Figure 2 .
Gene silencing efficiency was assessed based on thermodynamic RNA-RNA interaction between siRNA (guide strand) and mRNA (target gene) using minimum free energy of hybridization. Secondary structure prediction of mRNA-siRNA complex is crucial for effective RNAi using minimum free energy as a target of structural accuracy Bret et al., 2005 Mathews 2005. The free energy hybridization of the siRNA was calculated using the online server BIBISERV2-RNAHYBRID Tuschl et al., 2002. This tool follows a dynamic programming algorithm for RNA secondary structure prediction. The secondary structure of the 3 designed siRNAs along with the target mRNAs are depicted in Figure 3 .
The hybridization structure is used for further evaluation of various factors such as linearity of the mRNA-siRNA hybrid and GC content of siRNA. The linearity of the structure was evaluated based on the nucleotide in the loop structure region. The number of nucleotides in the loop region was inversely proportional to free energy and RNAi activity. The GC content of a siRNA is one of the essential parameters that might correlate with siRNA functionality. There is a negative correlation between target site accessibility and GC content. It is preferable to pick siRNAs with low GC content. Indeed, the GC content of the 3 siRNAs was in the range of 47-52%. The efficiency of silencing also depends on low GC content of siRNA. The best fitted siRNA (siRNA1) had 47.62% GC content. Thus, GC content is inversely proportional to RNAi mechanism Amarzguioui et al., 2004Reynolds et al., 2004.
In present study, three potential siRNA molecules were designed for silencing target TRPC5 mRNA. The best fitted siRNA was found to be siRNA1; it fulfilled all the necessary parameters, therefore supporting more effective binding of siRNA with target mRNA. Even though RNAi uses double stranded RNA in the form of siRNA or miRNA, siRNA is relatively superior than miRNA. This is due to the sequence specificity of siRNA and its delivery mechanism into the cell. siRNA mediated therapeutics have been efficient against metabolic disorders of liver and hypercholesterolemia in other studies Czech et al., 2011. Hence, RNAi may have great potential for treatment of challenging or incurable conditions (e.g. glomerular filtration malfunction).
Conclusion
RNAi technology is the leading strategy and therapy for various incurable diseases and genetic disorders. The compatibility of siRNA with the target gene can be assessed using bioinformatics tools. In this study, siRNA1 had the least minimum free energy of hybridization (i.e. -28.2 kcal/mol), low GC content of 47.62%, and high linearity with minimal h-b index and loop structure. It is predicted to be the most efficient towards the TRPC5 gene silencing. Hence, siRNAs can be used as a therapeutic drug to act as an important inhibitor to suppress gene expression. Thus, siRNAs represent a potentially effective therapeutic strategy for treating glomerular filtration malfunction. Further in vivo investigations are warranted to confirm their efficacy.
Author Contribution
All authors were equally contributed to the study design, bioinformatics analysis, drafting of the manuscript and approved the manuscript for publication.
References
-
M.
Amarzguioui,
H.
Prydz.
An algorithm for selection of functional siRNA sequences. Biochemical and Biophysical Research Communications.
2004;
316(4)
:
1050-1058
.
View Article Google Scholar -
N. S.
Anavekar,
D. J.
Gans,
T.
Berl,
R. D.
Rohde,
W.
Cooper,
A.
Bhaumik,
M. A.
Pfeffer.
Predictors of cardiovascular events in patients with type 2 diabetic nephropathy and hypertension: A case for albuminuria. Kidney International. Supplement.
2004;
92(92)
:
S50-S55
.
-
E.
Bernstein,
A. A.
Caudy,
S. M.
Hammond,
G. J.
Hannon.
Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature.
2001;
409(6818)
:
363-366
.
View Article PubMed Google Scholar -
G.
Berrut,
B.
Bouhanick,
P.
Fabbri,
G.
Guilloteau,
F.
Bled,
J. J.
Le Jeune,
M.
Marre.
Microalbuminuria as a predictor of a drop in glomerular filtration rate in subjects with non-insulin-dependent diabetes mellitus and hypertension. Clinical Nephrology.
1997;
48(2)
:
92-9
.
PubMed Google Scholar -
SE
Bret,
Soifer
Harris S.,
Bowers
Chauncey,
J. Rossi.
John.
siRNA target site secondary structure predictions using local stable substructures. Nucleic Acid Res.
2005;
33(3)
:
e30
.
View Article Google Scholar -
T. R.
Brummelkamp,
R.
Bernards,
R.
Agami.
A system for stable expression of short interfering RNAs in mammalian cells. Science.
2002;
296(5567)
:
550-553
.
View Article PubMed Google Scholar -
M. P.
Czech,
M.
Aouadi,
G. J.
Tesz.
RNAi-based therapeutic strategies for metabolic disease. Nature Reviews. Endocrinology.
2011;
7(8)
:
473-484
.
-
S. M.
Elbashir,
W.
Lendeckel,
T.
Tuschl.
RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Development.
2001;
15(2)
:
188-200
.
View Article PubMed Google Scholar -
M. G.
Farquhar.
The glomerular basement membrane: Not gone, just forgotten. The Journal of Clinical Investigation.
2006;
116(8)
:
2090-2093
.
View Article PubMed Google Scholar -
C.
Faul,
M.
Donnelly,
S.
Merscher-Gomez,
Y. H.
Chang,
S.
Franz,
J.
Delfgaauw,
P.
Mundel.
The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nature Medicine.
2008;
14(9)
:
931-938
.
View Article PubMed Google Scholar -
H. C.
Gerstein,
J. F.
Mann,
Q.
Yi,
B.
Zinman,
S. F.
Dinneen,
B.
Hoogwerf,
S.
Yusuf,
the HOPE Study Investigators.
Albuminuria and risk of cardiovascular events, death, and heart failure in diabetic and nondiabetic individuals. Journal of the American Medical Association.
2001;
286(4)
:
421-426
.
View Article PubMed Google Scholar -
S. M.
Hammond,
E.
Bernstein,
D.
Beach,
G. J.
Hannon.
An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature.
2000;
404(6775)
:
293-296
.
View Article PubMed Google Scholar -
B.
Haraldsson,
J.
Nyström,
W. M.
Deen.
Properties of the glomerular barrier and mechanisms of proteinuria. Physiological Reviews.
2008;
88(2)
:
451-487
.
View Article PubMed Google Scholar -
A. L.
Jackson,
P. S.
Linsley.
Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature Reviews. Drug Discovery.
2010;
9(1)
:
57-67
.
-
W. F.
Keane,
B. M.
Brenner,
D.
de Zeeuw,
J. P.
Grunfeld,
J.
McGill,
W. E.
Mitch,
R.
Toto,
the RENAAL Study Investigators..
The risk of developing end-stage renal disease in patients with type 2 diabetes and nephropathy: The RENAAL study. Kidney International.
2003;
63(4)
:
1499-1507
.
View Article PubMed Google Scholar -
H.
Ma,
A.
Togawa,
K.
Soda,
J.
Zhang,
S.
Lee,
M.
Ma,
S.
Ishibe.
Inhibition of podocyte FAK protects against proteinuria and foot process effacement. Journal of the American Society of Nephrology.
2010;
21(7)
:
1145-1156
.
View Article PubMed Google Scholar -
Y.
Ma,
C. Y.
Chan,
M. L.
He.
RNA interference and antiviral therapy. World Journal of Gastroenterology.
2007;
13(39)
:
5169-5179
.
View Article PubMed Google Scholar -
D. H.
Mathews.
Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics (Oxford.
2005;
England)
:
21(10)
.
View Article PubMed Google Scholar -
C. E.
Mogensen.
Microalbuminuria predicts clinical proteinuria and early mortality in maturity-onset diabetes. The New England Journal of Medicine.
1984;
310(6)
:
356-360
.
View Article PubMed Google Scholar -
C.
Montell.
TRP channels in Drosophila photoreceptor cells. The Journal of Physiology.
2005;
567(Pt 1)
:
45-51
.
View Article PubMed Google Scholar -
I. S.
Ramsey,
M.
Delling,
D. E.
Clapham.
An introduction to TRP channels. Annual Review of Physiology.
2006;
68(1)
:
619-647
.
View Article PubMed Google Scholar -
M.
Rehmsmeier,
P.
Steffen,
M.
Hochsmann,
R.
Giegerich.
Fast and effective prediction of microRNA/target duplexes. RNA (New York, N.Y.).
2004;
10(10)
:
1507-1517
.
-
A.
Reynolds,
D.
Leake,
Q.
Boese,
S.
Scaringe,
W. S.
Marshall,
A.
Khvorova.
Rational siRNA design for RNA interference. Nature Biotechnology.
2004;
22(3)
:
326-330
.
View Article PubMed Google Scholar -
A.
Riccio,
Y.
Li,
J.
Moon,
K. S.
Kim,
K. S.
Smith,
U.
Rudolph,
D. E.
Clapham.
Essential role for TRPC5 in amygdala function and fear-related behavior. Cell.
2009;
137(4)
:
761-772
.
View Article PubMed Google Scholar -
S. M.
Elbashir,
J.
Harborth,
W.
Lendeckel,
A.
Yalcin,
K.
Weber,
T.
Tuschl.
Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature.
2001;
411(6836)
:
494-498
.
View Article PubMed Google Scholar -
schaldecker
Thomas,
Kim
Sookyung,
Tarabanis
Constantine,
Tian
Dequan,
Hakroush
Samy,
Castonguay
Philip,
Ahn
Wooin,
Wallentin
Hanna,
Heid
Hans,
R. Hopkins
Corey,
W. Lindsley
Craig,
Riccio
Antonio,
Buvall
Lisa,
Weins
Astrid,
Greka
Anna.
Inhibition of the TRPC5 ion channel protects the kidney filter. J Clin Invest.
2013;
123(12)
:
5298-5309
.
-
D.
Tian,
S. M.
Jacobo,
D.
Billing,
A.
Rozkalne,
S. D.
Gage,
T.
Anagnostou,
A.
Greka.
Antagonistic regulation of actin dynamics and cell motility by TRPC5 and TRPC6 channels. Science Signaling.
2010;
3(145)
:
ra77
.
View Article PubMed Google Scholar -
A.
Togawa,
J.
Miyoshi,
H.
Ishizaki,
M.
Tanaka,
A.
Takakura,
H.
Nishioka,
Y.
Takai.
Progressive impairment of kidneys and reproductive organs in mice lacking Rho GDIalpha. Oncogene.
1999;
18(39)
:
5373-5380
.
View Article PubMed Google Scholar -
T.
Tuschl,
A.
Borkhardt.
Small interfering RNAs: Arevolutionary tool for the analysis of genefunction and genetherapy. Molecular Interventions.
2002;
2(3)
:
158-167.
.
View Article PubMed Google Scholar -
S. Z.
Xu,
P.
Sukumar,
F.
Zeng,
J.
Li,
A.
Jairaman,
A.
English,
D. J.
Beech.
TRPC channel activation by extracellular thioredoxin. Nature.
2008;
451(7174)
:
69-72
.
View Article PubMed Google Scholar -
E.
Yanagida-Asanuma,
K.
Asanuma,
K.
Kim,
M.
Donnelly,
H.
Young Choi,
J.
Hyung Chang,
P.
Mundel.
Synaptopodin protects against proteinuria by disrupting Cdc42:IRSp53:Mena signaling complexes in kidney podocytes. American Journal of Pathology.
2007;
171(2)
:
415-427
.
View Article PubMed Google Scholar -
P.D.
Zamore,
T.
Tuschl,
P.A.
Sharp,
D.P.
Bartel.
RNAi:Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell.
2000;
101(1)
:
25-33
.
View Article PubMed Google Scholar
Comments
Downloads
Article Details
Volume & Issue : Vol 5 No 1 (2018)
Page No.: 1911-1922
Published on: 2018-01-18
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 6814 times
- Download PDF downloaded - 2100 times
- View Article downloaded - 0 times
 Biomedpress
Biomedpress