Optimizing a multiplex high resolution melting curve to diagnose G6PD deficiency based on viangchan and canton mutations
Abstract
Glucose-6-phosphate dehydrogenase (G6PD) deficiency which is caused by mutation on G6PD gene is the most common enzyme disorder in human. There have been 184 discovered mutations among which Viangchan [Val291Met] and Canton mutation [Arg459Leu] are the most common variants in Vietnamese. Due to the severity of this disease, several methods have been devised for diagnostics. However, time-consuming, low sensitivity and expensiveness are major problems of those techniques. Recently, High Resolution Melting (HRM) has been developed and proven to be an effective method for DNA genotyping, mutation scanning and sequence matching. Hence, in this study a multiplex HRM has been developed aiming at detecting these two mutations concurrently. At first, a singleplex HRM was designed for each mutation. Then, conditions for these singleplex assay were combined and optimized again in order to get the optimal condition for multiplex HRM. Although this method showed a promising potential with a high accuracy, sensitivity, and specificity, it is impractial to continue developing this method because of the lack of controls. However, the optimized singleplex PCR-HRM in this study still can be used for single mutation detection and serve as the background to develop PCR-HRM for other G6PD mutations in Vietnamese-Kinh population.
Introduction
In Viet Nam, scientists have been studying about molecular mechanism of the G6PD deficient since 2000 (Dao et al., 2000) and have found common mutations in some provinces and populations such as Kinh and others populations in Lam Dong Matsuoka et al., 2007, Kinh in Ho Chi Minh city, and S’tieng in Binh Phuoc Hue et al., 2013bHue et al., 2009. In 2013, two causative mutations of the G6PD deficiency in Kinh population have been determined including Viangchan (871G>A) and Canton (1376G>T) Hue et al., 2013aHue et al., 2013c. These two missense mutations have been screened and occurred with high frequencies in the G6PD deficient patients (31.57% and 26.3%) according to the study in Lam Dong province Matsuoka et al., 2007 and 43.33% and 26.67% according to the study in Ho Chi Minh Hue et al., 2013b. However, up to now, there is not any molecular method applied for diagnosing the G6PD deficiency in Viet Nam. Therefore, this study is conducted to develop a successful HRM method to detect two causative G6PD mutations in Vietnamese-Kinh population.
Methods
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is the most common enzymatic disorder affecting human beings by influencing on red blood cells (RBCs). G6PD can be found throughout the body but its role is more crucial in RBCs then in others as it is the only NADPH-producing enzyme activated during oxidative stress. In the cells, G6PD participates in the pentose phosphate pathway and catalyzes its first step. This enzyme converts Glucose-6-phosphate into to 6-phospho D-gluconolactone and turns NADP to NADPH at the same time. NADPH is a necessary substance to protect the cell especially RBCs and its haemoglobin from oxidative stress created by reactive oxidation species (ROSs) such as fava beans, antimalaria drug or fava plant Beutler, 1994. Thus, in the condition of G6PD deficiency, when oxidative stress happens, red blood cells undergo the premature rupture and breakdown, a process called hemolysis, faster than the body can replace them. This loss of red blood cells causes the signs and symptoms of hemolytic anemia, which is a characteristic feature of glucose-6-phosphate dehydrogenase deficiency Noori-Daloii et al., 2006. According to WHO, based on the residual enzymatic activity and clinical manifestation, G6PD deficiency can be classified into five classes Frank, 2005, from severe enzyme deficiency to increased enzymatic activity. As reported by many studies on the G6PD deficiency, the cause of this disease is mutations on G6PD gene that lead to changes in the structure of G6PD enzyme. Today, more than 184 mutations Minucci. A, 2012 have been found and most of them (159 mutations) are point mutations which result in the amino acid substitutions. The mutations occurring inside the active site or in the promoter region, as well as the de no vo mutations (causing sporadic variants) have also been identified but they are quite rare. Besides,the large deletions and frame-shift mutations that lead to the complete lacking of the G6PD functionality are not observedAnna L. Peters and Noorden, 2009Minucci et al., 2009Philip J Mason, 2007. Therefore, most of the G6PD deficient cases can be diagnosed by detecting the mutations occurring on the G6PD gene.
From these results, scientists are now focusing on creating the molecular methods that support to biochemical methods for increasing the accuracy and the specificity of the tests. Depending on each population, different methods have been developed to determine the causative mutations in this population. For example, in Africa where the A- mutation (202G>A, 376A>G) occupies more than 90% deficient cases, the molecular test have been successfully developed and applied by the Arup Laboratory in Utal university Laboratories, 2015. In addition, the molecular tests are also applied to detect Mediterranean mutation (563C>T), a common in the Southern Europe and Viangchan mutation (871G>A), the main kind in Lao and Cambodian widely Joly, 2010. In China where has the close genetic interaction with Viet Nam, nine mutations (A95G, G392T, G487A, A493G, C592T, C1024T, C1360T, G1376T, G1388A) occupying more than 90% of G6PD deficient cases have been determined Jiang et al., 2006Yan et al., 2006. In these mutations, the Canton mutation (G1376T) and Kaiping mutation (G1388A)- two missense mutations in type 2 – occupying 53.8% have been successfully diagnosed by the HRM analysis (high resolution melting analysis) Yan et al., 2010. Recently, this method has been developed and allows the scientists quickly detect and discriminate different genotypes of different mutations before performing the sequencing Reed et al., 2007.
Sample preparation
Sample collection
Dried blood samples of infants with consent forms were collected from Tu Du Maternity hospital – Ho Chi Minh City – Vietnam since 2009 to 2012. The blood spots samples used were air-dried for 4 hours at room temperature and stored in plastic bags to protect them from moisture, light, heat and contact with other materials. Then, these samples were screened for G6PD deficiency by the Neonatal G6PD Screening Assay (Zentech, Belgium) and genotyped for Canton and Viangchan mutation by ARMS-PCR method. Then, several samples with genotypes of YG, GA, YA (Viangchan mutation) and of YG, GT, YT (Canton mutation) have been sequenced to confirm ARMSPCR result and were used as control in real-time PCR reaction (in High Resolution Melting Analysis method).
DNA extraction
DNA were extracted by QIAmp DNA blood mini kit from these dried blood spots. The quantity and quality were then verified by NanoDrop 1000 spectrophotometer (Thermo Scientific, USA). And only DNA solutions with OD value of A260/A280 in range of 1.7 to 2.0 were used.
Optimizing singleplex PCR-HRM reaction for each mutation
Primer design
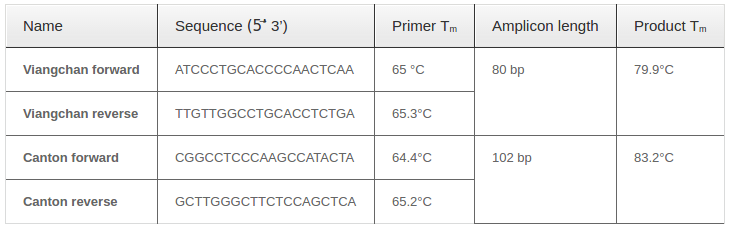
Primers used for PCR-HRM reaction were designed by Primer3plus website Untergasser et al., 2012 with several criteria, including: 80-to-150bp PCR product, 20-to-27bp primer, and melting temperature between 60-65°C. The specificity of primer pairs was tested using BLAST tool provided by NCBI website Altschul et al., 1990.
Predicted amplicons’ melting curve of selected primers were also checked using uMELT website. Optimal concentration of MgCl2and DMSO was also predicted through this website by adjusting concentration until three melting curves of various genotypes could be distinguished clearly.
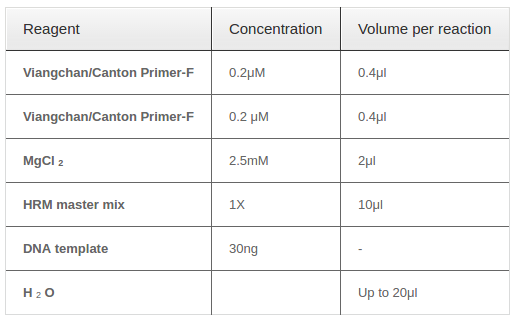
PCR reagents optimization
The annealing temperature was determined by running numerous PCR reactions at different annealing temperature within the range of 58°C to 68°C, with 2°C of increment. In addition, optimal MgCl2concentration was tested again because the concentration given by uMELT web tool was only a predicted one. A range of MgCl2concentrations surrounding the predicted one which was from 1mM to 3mM was tested to select the optimal one.
Melting curve prediction
The predicted melting curve and melting peak could be distinguished clearly with 2mM MgCl2, proving that the primer pair for Viangchan was suitable for real-time PCR with HRM analysis. The specificity of primer was checked on NCBI website.
Test validation for each mutation
The optimized PCR-HRM for each mutation detection was validated by genotyping a large number of samples. For Viangchan mutation, the test included 50 samples already genotyped by ARMS-PCR method and 273 unknown-genotype samples from OUCRU to compare their accuracy. These 273 samples were also used for Canton-test validation. Based on melting curve, different plot, and melting peak generated by HRM analysis the validation of the test can be concluded.
Combine two singleplex PCR into multiplex PCR
After checking capability of each primer set, two set of primers were added to the same PCR reaction aiming at developing multiplex PCR detecting these SNP. The results were then analyzed based on HRM result, specifically melting curves and melting peaks.
Results
Primer design and melting curve prediction
Using the Primer3plus, four sets of primers were designed to produce a PCR product suitable for the selected genotyping method. Each includes one reverse and one forward primer. Primers and amplicon characteristics are summarized in Table 1 Table 1 and predicted melting curves and melting peaks are shown in Figure 1 Figure 2 .
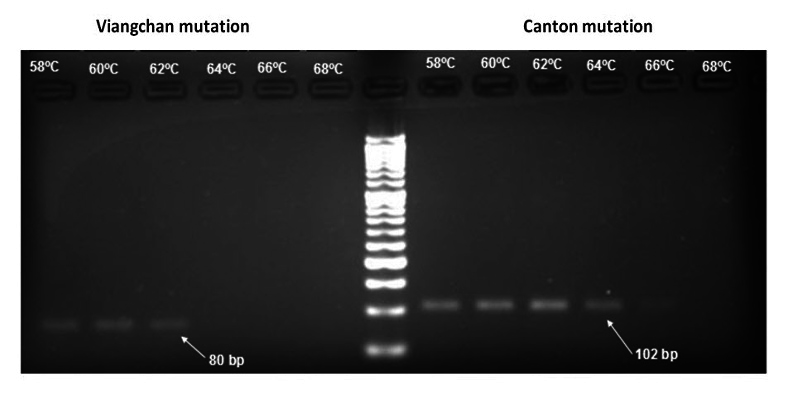
Annealing temperature optimization
After running PCR in a gradient of temperature condition, the results showed that whilst primers of Viangchan mutation worked best at 60°C, the optimal annealing temperature for Canton was 62°C because at those temperatures the bands were brightest and clearest which indicated the reactions ran efficiently Figure 3 . However, with the purpose of developing multiplex PCR-HRM, 62°C was chosen as the suitable one.
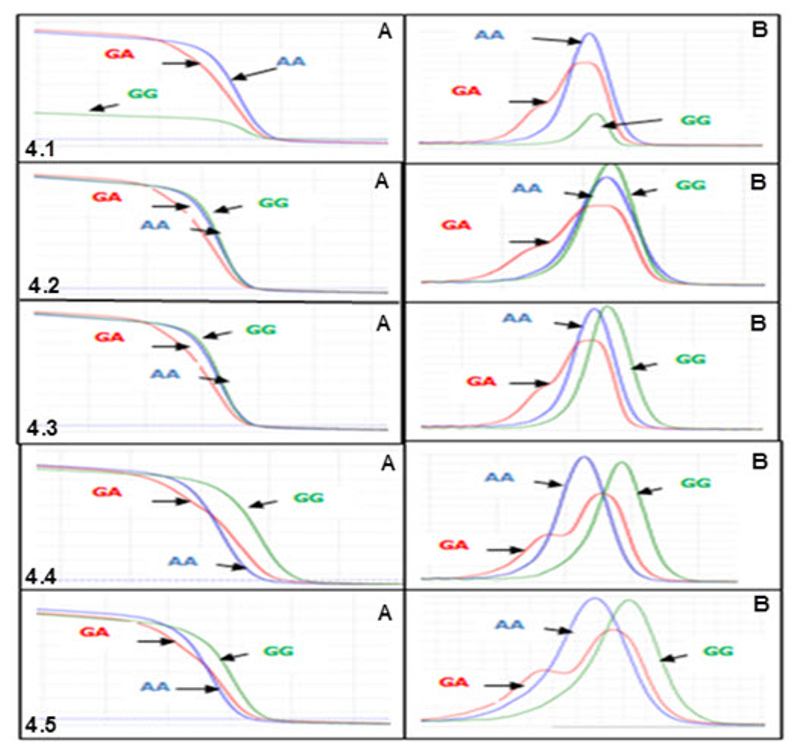
MgCl2 concentration optimization
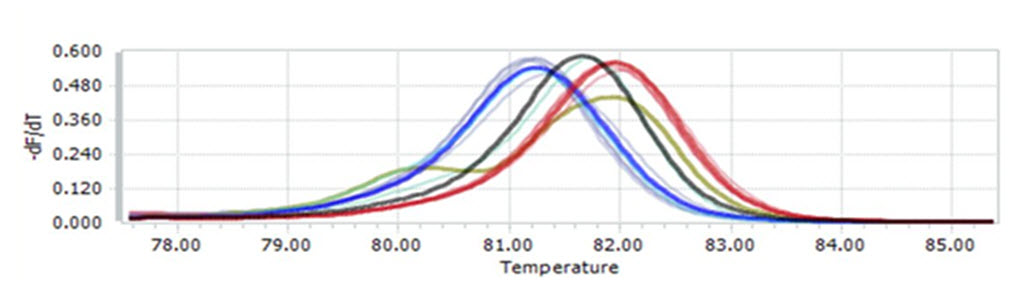
Because MgCl2concentration had significant effect on melting curve of PCR products, PCR-HRM reactions was run with different MgCl2 concentrations to determine the optimal one. The result revealed that at concentration of 1.0mM, not only the melting curve of GG was lower than two other curves (Fig 4.1A) but also the minor melting peak of heterozygous genotype did not appeared clearly (Fig 4.1B) . This indicated that PCR- HRM reaction did not run effectively at this concentration. When the concentration grew to 1.5mM and 2mM, the PCRHRM reaction was likely to worked better because three melting curves were generated appropriately. However, for HRM analysis these concentrations were not the best choice because curves of two homologous genotypes (AA, GG) were clustered (Fig. 4.2A , 4.3A ) .
This may lead to missed calling two genotypes when analyzing many samples. Among tested concentrations, 2.5mM and 3.0mM of MgCl2 seemed to give good result for HRM analysis because at these concentrations the melting curves as well as melting peaks showed similar, clear, and correct shapes (Fig. 4.4, 4.5) . Nonetheless, the melting curves at 2.5mM showed better discrimination, especially between two homozygous-genotype curves. Thus, MgCl2 2.5mM was chosen as the optimal concentration for PCRHRM.
In summary, after optimizing all crucial components for a PCR-HRM analysis, the optimum conditions to detect Viangchan and Canton mutation were identified Table 2 and prepared for further application.
PCR-HRM result of control samples
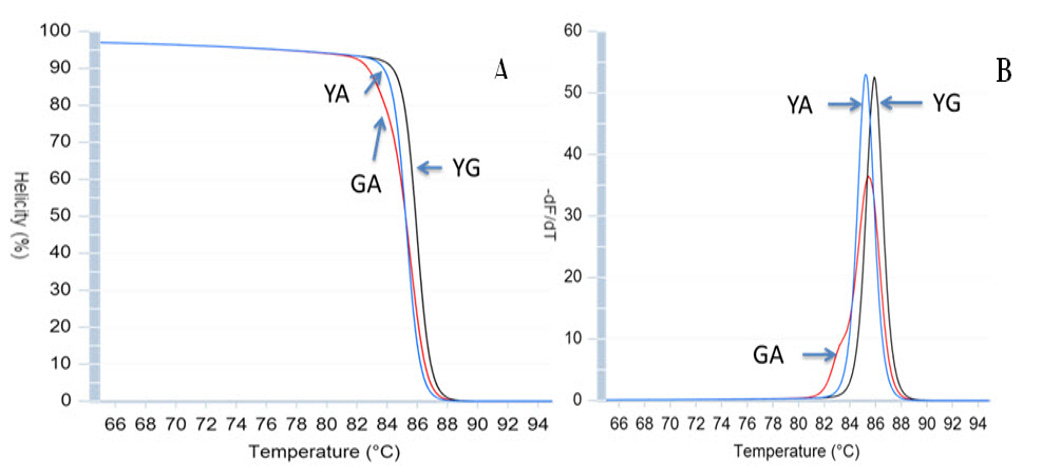
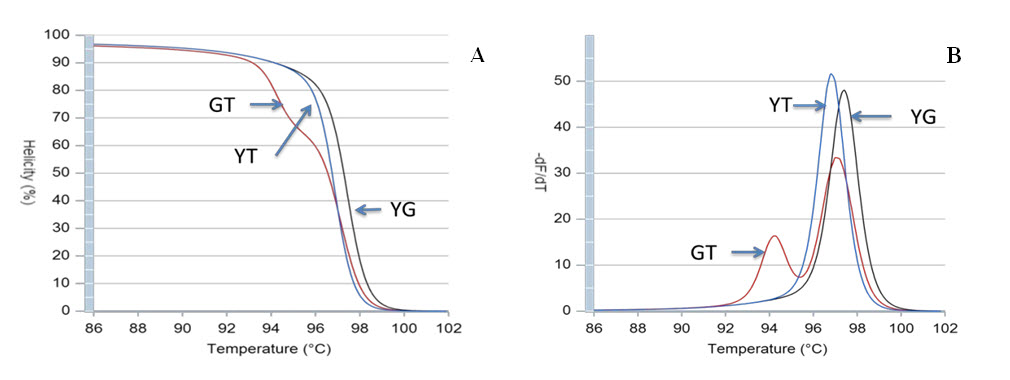
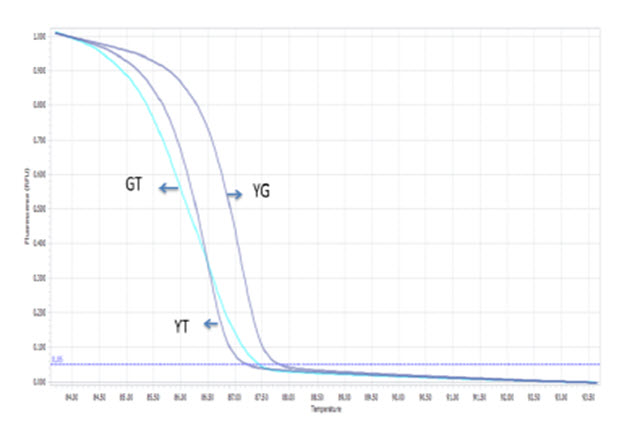
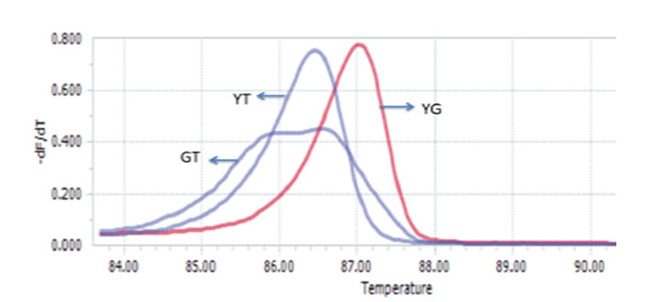
The optimized PCR protocol was examined by genotyping again sequenced samples of Viangchan or Canton mutations. The results were analyzed based on melting cure, melting peak and different plot. It can be easily seen that with these optimal conditions all melting curves as well as melting peaks were generated clearly. Although melting peaks of Viangchan heterozygous sample seemed better than those of Canton sample, their melting curves, especially, two curves representing for hemizygous wild type and hemizygous mutant samples can obtain a fine discrimination ( Figure 5A , Figure 5B , Figure 6A , Figure 6B ). This indicated that under these conditions PCR-HRM worked effectively.
Based on these patterns, genotype of a sample can be identified depending on which control peak ( Figure 5A Figure 6A ) or curve ( Figure 5B Figure 6B ) or plot ( Figure 5C , Figure 6C ) they cluster around. Hence, there would bethree methods to determine the genotype of a sample through HRM analysis. This leads to an increase in accuracy of genotype determination.
Test validation for singleplex PCR-HRM method
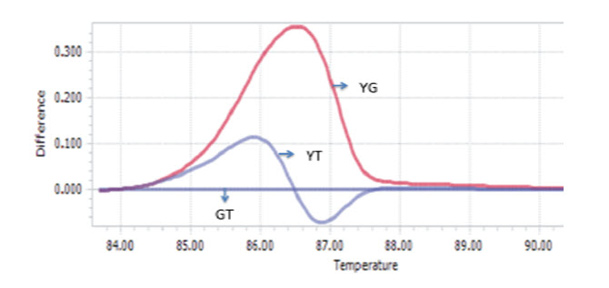
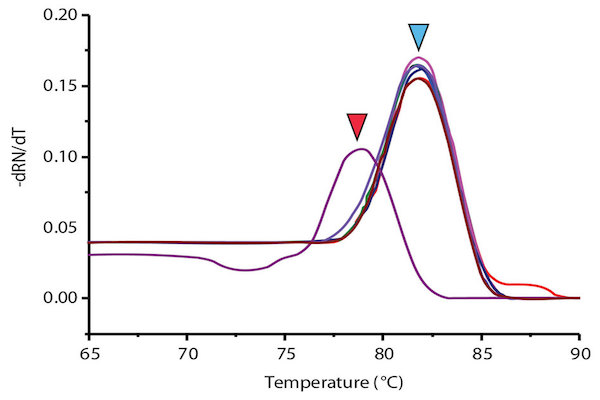
When applying singleplex PCR-HRM in regenotyping 50 samples with Viangchan mutation (from Tu Du hospital), the result of 47 samples showed a consistency between PCR-HRM and ARMS-PCR method which was used for genotyping previously. The remaining three samples had different results between these two methods. So, they were sequenced to determine the exact genotypes. The sequencing results indicated that PCR-HRM method gave correct genotypes. However, one problem could occur by using PCR-HRM is shifted melting peak which mean that instead of clustering around control melting-peak, the peak lay between peaks of two hemizygous samples. This would lead to difficulty in genotype determination Figure 7 . To solve this issue, DNA from such sample was extracted from dried blood, and then genotyped again. The melting peak of this sample now clustered with the control of wild type, similar result with ARM-PCR method.
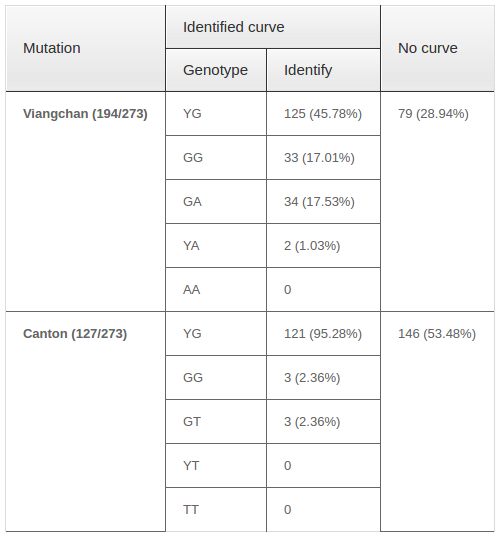
Viangchan and Canton mutation were then continued to be screened in 273 unknown-genotyped samples from Oxford University Clinical research Unit (OUCRU). The result showed that for Viangchan mutation, 71.06% (194/273) of samples were genotyped successfully while the rest did not generate any melting curve. Similar situation was found for Canton mutation. However, the percentage of samples whose genotype can be identified was lower, only 46.52% (127/273). The genotyping result was summarized in Table 3 .
Multiplex PCR-HRM
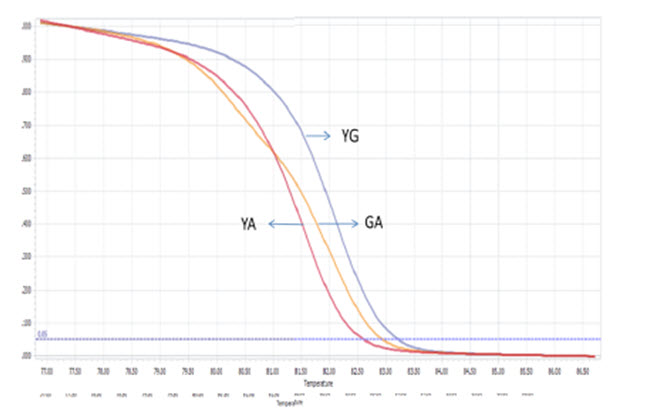
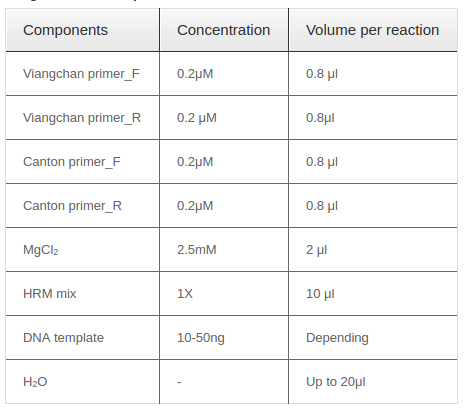
Two singleplex PCR reactions were then combined into one in order to detect two mutations at the same time. Reagents needed for one multiplex PCR-HRM were listed in Table 4 with required concentration and corresponding volume. Because of different amplicon sizes, PCR products of two mutations would had distinct melting temperatures. Thus, multiplex reaction outcome should had a melting curve with two shoulders, one represented for Viangchan mutation and the other for Canton. Besides, the shoulder for Viangchan mutation would appeared earlier due to lower product meltingtemperature ( Table 1 ), as showed in Figure 8 . The results of multiplex PCR-HRM can also be evaluated with melting peak.
Using the same controls with singlex, multiplex PCRHRM was optimized. Theoretically, there should be nine controls for multiplex PCR because of various combinations between three genotypes of two mutations. Nonetheless, by reasons of the limitation of samples and rareness of genotype variants, there were only 4 controls for optimization.
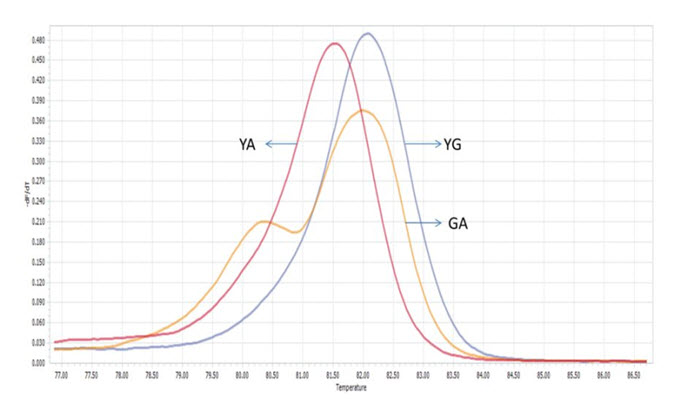
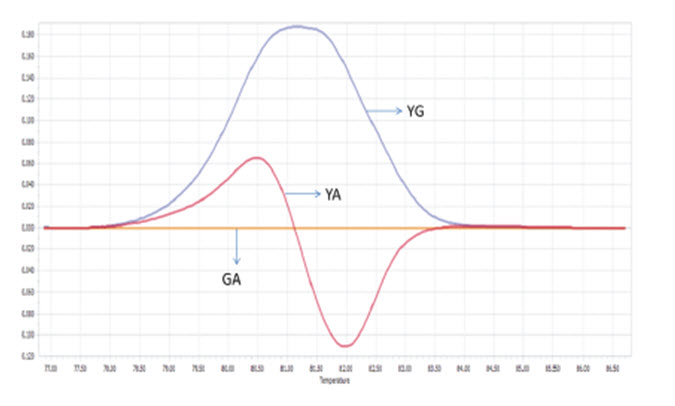
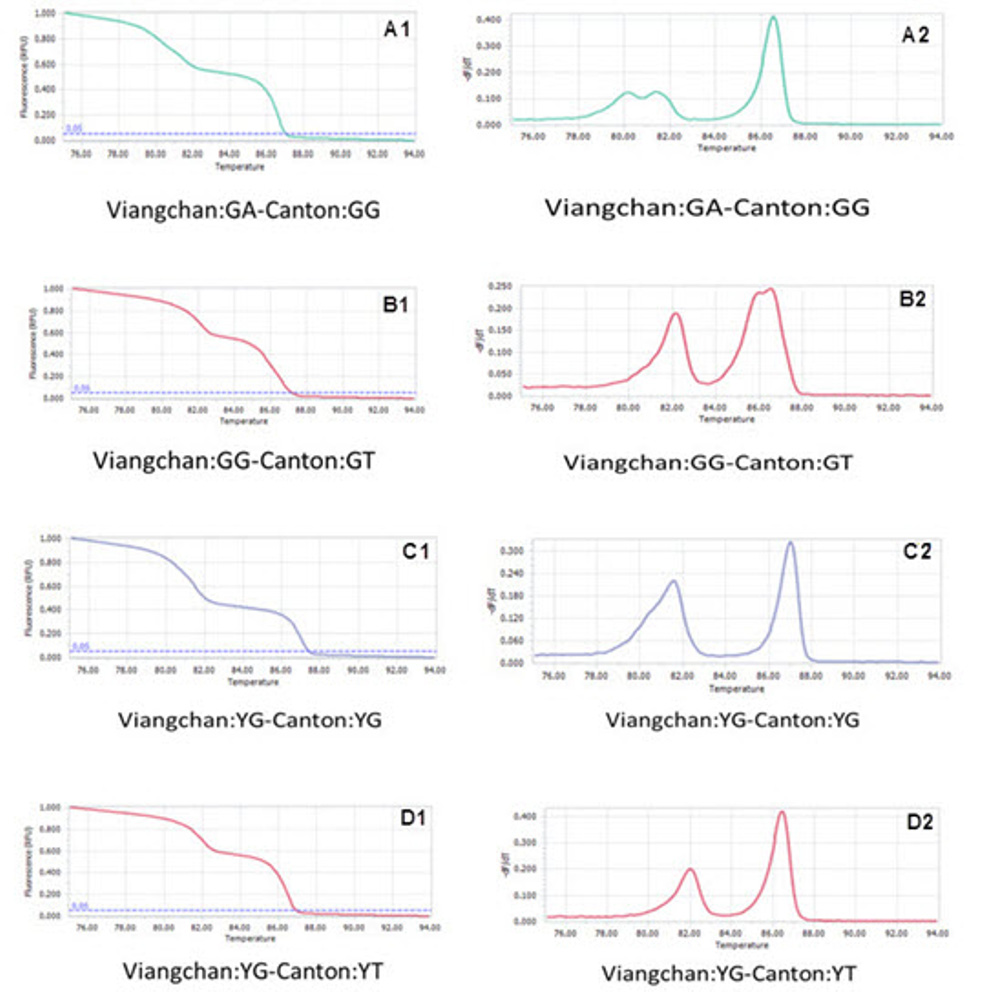
Among 50 samples which had already been genotyped by ARMS-PCR and confirmed by sequencing, four samples with both Viangchan and Canton mutation were used as controls for multiplex PCR-HRM. Genotype combinations of these samples are: hemizygous wild type for both mutations, heterozygous Viangchan and homozygous wild-type Canton, homozygouswild-type Viangchan and heterozygous Canton, and hemizygous mutants for both mutations. Multiplex PCR-HRM reactions for four control samples were run separately in order to identify the curve shapes of each combination. As expected, two shoulders in melting curve which represent the presence of two mutations were shown. Nevertheless, the genotype identification became relatively more complicated because the difference in various genotypes’ melting curve shape of a mutation was not as clear as in singleplex (Fig. 9A1 , B1 , C1 , D1 ) . However, by analyzing melting peaks, various genotypes of each mutation as well as four genotypecombinations can be distinguished easier. For example, for the combination of heterozygous and homozygous genotype there were three melting peaks. Two of those were very closed to each other and quite separated from the other one (figure 9A2, B2) . Similar to singleplex PCR, these two closed peaks represented for heterozygous genotype while the “only one peak” indicated the homozygous genotype. Yet, the analysis of melting peak patterns only could not help to differentiate between combination of two hemizygous-genotype (Fig. 9C2 , D2 ) . In this case, melting temperature (Tm) would become a solution. First, due to a bigger amplicon, Canton mutation would have higher Tm thanViangchan mutation. Specificially, Tm of Canton mutation was 87°C while Viangchan mutation had Tm of 81.5°C.Then, in figure 9 C2 and D2 when genotype of Canton mutation changed from YG to YT, its melting temperature (Tm) also decreased from 87°C to around 86.5°C.
Therefore, multiplex PCR-HRM can help to determine these four genotype combinations. In the other words, if an unknown sample has a similar curve-shape with one of these four controls, the genotype of Viangchan and Canton mutation could be identified at the same time.
Discussion
G6PD deficiency is an enzymatic disorder caused by mutations on G6PD gene. Although this disease can lead to mild or severe hemolysis, patient still can live a normal life if they have suitable drug and diet. In order to help them, numerous molecular methods for G6PD-deficiency diagnosis have been developed until now. In this study, a multiplex PCR-HRM has been devised to detect Viangchan and Canton mutation due to their high frequency in Vietnamese population. When applying singleplex PCR-HRM to genotype these mutations, many strengths of this method have been found. It is a simple and fast work flow technique. It does not require any additional instrumentation or steps besides PCR preparation and a PCR machine capable of detecting the dye. In addition, toxic chemical like EtBr or post-PCR extrinsic error can be eliminated since running gel electrophoresis is no longer necessary for this method. Especially, HRM has high sensitivity and accuracy. This can obviously be seen when comparing Viangchan genotyping result between this method with ARMS-PCR. It can be found that among 50 genotyped samples, ARMS-PCR gave 47/50 correct genotypes while HRM gave exactly 50/50 correct ones. This has proven that PCR-HRM is a promising method for genotyping. Nevertheless, this high sensitivity can also become its weakness. First of all, because of this characteristic, it can get contamination easily in a run. Second, because HRM is highly sensitive, only small difference in DNA quality can also affect amplification process remarkably resulting in shifting melting curve. For example, in this study, when a DNA sample which was stored for more than two year was used for genotyping, it gave a shifted or unidentified melting curved. However, when this reaction was performed again with new extracted DNA, its melting curve was identified. From this experiment, it can also be concluded that DNA quality had remarkable effect on the result of HRM analysis.
Additionally, the primer design step should be pay more attention because the generation of effective amplicon is vital for this method. Specificially, the amplicon length may have significant influence on the sensitivity of HRM method. For a short amplicons, if there is a presence of a single base difference, melting temperature (Tm) among different genotypes is noticeable. This allows good discrimination among them. However, when the amplicon size increases, the Tm difference decreases resulting in difficulty in genotypes differentiation Erali et al., 2008Krypuy et al., 2006Liew et al., 2004. For this project, the amplicon size were 80bp and 102bp which were in the optimal range for HRM analysis. Moreover, short amplicon also limits the number of mutations presence on it which may affect the melting curve pattern. It was reported that a few G6PD variants have also occurred in close proximity of these mutations (Available from: Ensembl 2014 Database; http://www.ensembl.org, Sep 28, 2015) but their frequencies are insignificant compared to Viangchan and Canton mutation .
In this study, even though a background singleplex PCR-HRM for each mutation was designed successfully, the development of multiplex PCR-HRM still has had few limitations. Firstly, though four melting curve patterns were identified, they were identified separately. Basically, one assay with four controls needs to be run so that various melting curve patterns can be compared and the ability to distinguish between different genotype combinations based solely on melting curve can be checked. This is necessary because it would help to improve the process of result analysis. Secondly, for this method, the genotype of a sample can only be determined if its melting curve is closely parallel with control’s melting curve. So, in order to fully develop this method and make it fucntion a set of nine controls which are nine genotype-combinations of two mutations is needed. This lack of controls is critical because it may lead to result mis-reading. Hence, to continue developing this method, other five missing genotype-combination, or control, should be created by mixing amplicons having different genotypes together. For instance, a heterozygous genotype combination of both mutations can be made by performing PCR for two samples each of which contains heterozygous Viangchan and heterozygous Canton and mixing products of two PCR reactions together.
Conclusion
In this study, singleplex PCR- HRM for detection of Viangchan and Canton mutations in G6PD gene has been performed relatively successful. Multiplex PCRHRM for detection of these mutations has also been designed although this performance was unsuccessful. With four controls available, four genotype combinations of these mutations can be identified. This also means that the genotype of each mutation can be determined immediately with only one reaction. Hence, in comparison with tranditional methods, this method shows number of advantages in numerous aspects. It is more economic efficiency, time saving, sensitivity, and specificity than other methods such as realtime PCR, Amplificationrefractory mutation system- PCR (ARMS- PCR), or even singleplex PCR- HRM. Nonetheless, the method also has it own disadvantages. Due to the high sensitivity of this method, it is easy to get contamination in a run. Also, quantity and quality of DNA input should be checked carefully because of their significant effect on HRM reaction which leads to shift in melting curve.
Although multiplex PCR-HRM has showed its potential in genotyping 2 mutations simultaneously, the study has encountered few difficulties among which limitation of control samples is the most considered one. In this study, only four out of nine controls presented. The other five controls are still needed to make this method function completely. This lead to an impracticality of continuing development of multiplex PCR-HRM. And multiplex PCR-HRM still cannot be applied into practice at this stage. However, singleplex PCR-HRM in this study can serve as the background to develop PCR-HRM for other G6PD mutations in Vietnamese-Kinh population. In the future, an identified amount of new dried blood samples will be used to performed again this singleplex PCR-HRM so as to determined exactly the eficiency of this method.
Abbreviations
G6PD: Glucose-6-Phosphate dehydrogenase; HRM: High Resolution Melting; RBC: Red blood cell: ROSs: reactive oxidation species; WHO: World Health Organization; ARMS-PCR: Amplification refractory mutation system-polymerase chain reaction; OUCRU: Oxford University Clinical research unit; PCR: Polymerase Chain reaction; SNP: Single nucleotide polymorphism.
References
-
S.F.
Altschul,
W.
Gish,
W.
Miller,
E.W.
Myers,
D.J.
Lipman.
Basic local alignment search tool. J Mol Biol.
1990;
215
:
403-410
.
-
Noorden
Anna L. Peters.
Glucose-6-phosphate Dehydrogenase Deficiency and Malaria: Cytochemical Detection of Heterozygous G6PD Deficiency in Women. Histochemistry & Cytochemistry 57.
2009;
11
:
1003-1011
.
-
E.
Beutler.
G6PD deficiency. Blood.
1994;
84
:
3613-3636
.
-
N.T.N.
Dao,
T.T.
Chinh,
T.T.
Tinh,
K.
Nishiyama,
T.
Shirakawa,
M.
Masafumi.
Analyzing some mutation variants of gene caused the G6PD- deficiency in Viet Nam. International Conference: Biotechnology-Environment and Non-Conventional Energy, Indian instittute of chemical technology.
2000
.
-
M.
Erali,
K.V.
Voelkerding,
C.T.
Wittwer.
High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol.
2008;
85
:
50-58
.
-
J.E.
Frank.
Diagnosis and management of G6PD deficiency. Am Fam Physician.
2005;
72
:
1277-1282
.
-
N.T.
Hue,
D.T.
Anh,
H.L.T.
Trinh,
P.N.
Hoang.
The Canton mutation in G6PD deficient Vietnamese. Wulfenia Journal.
2013a;
20
:
229-249
.
-
N.T.
Hue,
D.T.
Anh,
H.L.T.
Trinh,
P.N.
Hoang.
Common mutations in G6PD of Vietnamese-Kinh deficient patients. African Journal of Biotechnology.
2013b;
12
:
1318-1325
.
-
N.T.
Hue,
D.T.L.
Anh,
N.D.T.
Giang,
P.N.
Hoang.
The Viangchan-G6PD Mutation in Vietnamese-Kinh. Int J Hum Genet.
2013c;
13
:
85-92
.
-
N.T.
Hue,
J.P.
Charlieu,
T.T.
Chau,
N.
Day,
J.J.
Farrar,
T.T.
Hien,
S.J.
Dunstan.
Glucose-6-phosphate dehydrogenase (G6PD) mutations and haemoglobinuria syndrome in the Vietnamese population. alar J.
2009;
8
:
152
.
-
W.
Jiang,
G.
Yu,
P.
Liu,
Q.
Geng,
L.
Chen,
Q.
Lin,
X.
Ren,
W.
Ye,
Y.
He,
Y.
Guo.
Structure and function of glucose-6-phosphate dehydrogenase-deficient variants in Chinese population. Hum Genet.
2006;
119
:
463-478
.
-
P.
Joly.
Rapid genotyping of two common G6PD variants, African (A-) and Mediterranean, by high-resolution melting analysis. Clin Biochem 43.
2010;
1-2
:
193-197
.
-
M.
Krypuy,
G.M.
Newnham,
D.M.
Thomas,
M.
Conron,
A.
Dobrovic.
High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer.
2006;
6
:
295
.
-
A.
Laboratories.
Glucose-6-Phosphate Dehydrogenase (G6PD) 2 mutations. Laboratory test directory Department of pathology, University of Utah, U.o. Utah, ed. (http://ltd.aruplab.com/Tests/Pub/0051684. Cited 26 Nov 2015).
2015
.
-
M.
Liew,
R.
Pryor,
R.
Palais,
C.
Meadows,
M.
Erali,
E.
Lyon,
C.
Wittwer.
Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem.
2004;
50
:
1156-1164
.
-
H.
Matsuoka,
D.T.
Thuan,
H.
van Thien,
T.
Kanbe,
A.
Jalloh,
M.
Hirai,
M.
Arai,
N.T.
Dung,
F.
Kawamoto.
Seven different glucose-6-phosphate dehydrogenase variants including a new variant distributed in Lam Dong Province in southern Vietnam. Acta Med Okayama.
2007;
61
:
213-219
.
-
A.
Minucci,
B.
Giardina,
C.
Zuppi,
E.
Capoluongo.
Glucose-6-phosphate dehydrogenase laboratory assay: How, when, and why?. IUBMB Life.
2009;
61
:
27-34
.
-
M.K.
Minucci. A,
Zuppi. C
Hwang. M. J.
Glucose-6-phosphate dehydrogenase (G6PD) mutations database: Review of the “old” and update of the new mutations. Blood.
2012;
Cells
:
Molecules, and Diseases 48, 154-165
.
-
M.
Noori-Daloii,
S.
Hejazi,
A.
Yousefi,
S.
Mohammad Ganji,
S.
Soltani,
D.
Javadi,
M.
Sanati.
Identification of Mutations in G6PD Gene in Patients in Hormozgan Province of Iran. Islamic Republic of Iran.
2006;
17
:
313-316
.
-
J.M.B.
Philip J Mason,
Gilsanz
Florinda.
G6PD deficiency: the genotype-phenotype association. Blood Reviews.
2007;
21
:
267-283
.
-
G.H.
Reed,
J.O.
Kent,
C.T.
Wittwer.
Highresolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics.
2007;
8
:
597-608
.
-
A.
Untergasser,
I.
Cutcutache,
T.
Koressaar,
J.
Ye,
B.C.
Faircloth,
M.
Remm,
S.G.
Rozen.
Primer3-new capabilities and interfaces. Nucleic Acids Res.
2012;
40
:
e115
.
-
J.B.
Yan,
H.P.
Xu,
C.
Xiong,
Z.R.
Ren,
G.L.
Tian,
F.
Zeng,
S.Z.
Huang.
Rapid and reliable detection of glucose-6- phosphate dehydrogenase (G6PD) gene mutations in Han Chinese using high-resolution melting analysis. J Mol Diagn.
2010;
12
:
305-311
.
-
T.
Yan,
R.
Cai,
O.
Mo,
D.
Zhu,
H.
Ouyang,
L.
Huang,
M.
Zhao,
F.
Huang,
L.
Li,
X.
Liang.
Incidence and complete molecular characterization of glucose-6-phosphate dehydrogenase deficiency in the Guangxi Zhuang autonomous region of southern China: description of four novel mutations. Haematologica.
2006;
91
:
1321-1328
.
Comments
Downloads
Article Details
Volume & Issue : Vol 3 No 08 (2016)
Page No.: 757-769
Published on: 2016-08-30
Citations
Copyrights & License

This work is licensed under a Creative Commons Attribution 4.0 International License.
Search Panel
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Google Scholar
Pubmed
Search for this article in:
Google Scholar
Researchgate
- HTML viewed - 5715 times
- Download PDF downloaded - 2008 times
- View Article downloaded - 13 times
 Biomedpress
Biomedpress